| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
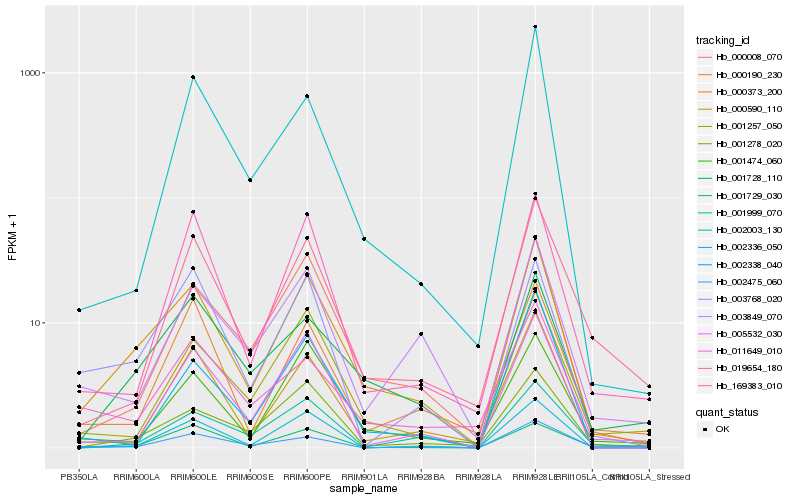
Hb_000373_200 |
0.0 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Populus euphratica] |
| 2 |
Hb_001728_110 |
0.1506596416 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF039-like [Jatropha curcas] |
| 3 |
Hb_000008_070 |
0.1512563173 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_001278_020 |
0.1575244883 |
- |
- |
sucrose synthase 6 [Hevea brasiliensis] |
| 5 |
Hb_001257_050 |
0.1652264064 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: protein YLS9 [Jatropha curcas] |
| 6 |
Hb_001474_060 |
0.1690695344 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 16 [Jatropha curcas] |
| 7 |
Hb_000590_110 |
0.169363456 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003768_020 |
0.171639696 |
- |
- |
PREDICTED: uncharacterized protein LOC105644157 [Jatropha curcas] |
| 9 |
Hb_005532_030 |
0.1723282627 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_019654_180 |
0.1758344423 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 11 |
Hb_011649_010 |
0.1766573734 |
transcription factor |
TF Family: Pseudo ARR-B |
sensory transduction histidine kinase, putative [Ricinus communis] |
| 12 |
Hb_002336_050 |
0.1773266289 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000190_230 |
0.1781123315 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 14 |
Hb_002475_060 |
0.1793103434 |
- |
- |
PREDICTED: potassium channel SKOR-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_001999_070 |
0.1809055634 |
- |
- |
PREDICTED: ammonium transporter 1 member 3 [Jatropha curcas] |
| 16 |
Hb_003849_070 |
0.1817217907 |
- |
- |
PREDICTED: uncharacterized protein LOC105644690 [Jatropha curcas] |
| 17 |
Hb_169383_010 |
0.1819914469 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |
| 18 |
Hb_002003_130 |
0.1820709124 |
- |
- |
hypothetical protein CISIN_1g027843mg [Citrus sinensis] |
| 19 |
Hb_002338_040 |
0.1824333007 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001729_030 |
0.1837838674 |
- |
- |
hypothetical protein VITISV_044457 [Vitis vinifera] |