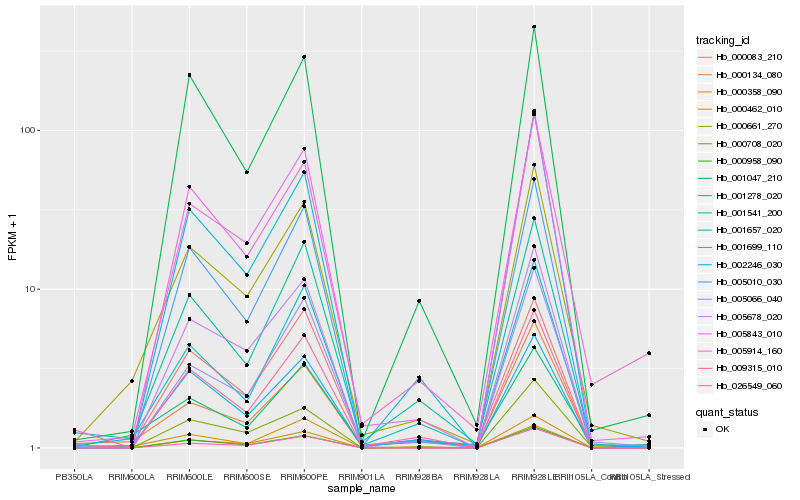
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000661_270 |
0.0 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_005843_010 |
0.0834342759 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 3 |
Hb_001278_020 |
0.0998457672 |
- |
- |
sucrose synthase 6 [Hevea brasiliensis] |
| 4 |
Hb_001541_200 |
0.1059763254 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 5 |
Hb_000134_080 |
0.1059893183 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 6 |
Hb_000708_020 |
0.1060036289 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 7 |
Hb_000083_210 |
0.1084849777 |
- |
- |
Major facilitator superfamily protein, putative [Theobroma cacao] |
| 8 |
Hb_001657_020 |
0.1100733648 |
- |
- |
heat shock family protein [Populus trichocarpa] |
| 9 |
Hb_005010_030 |
0.1101334939 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 1 [Jatropha curcas] |
| 10 |
Hb_005678_020 |
0.1120247454 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_026549_060 |
0.1125561417 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 12 |
Hb_000358_090 |
0.1134647787 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_005066_040 |
0.1134859574 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 14 |
Hb_002246_030 |
0.1143811475 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 15 |
Hb_005914_160 |
0.1151367473 |
- |
- |
PREDICTED: protein PROTON GRADIENT REGULATION 5, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000958_090 |
0.1167209804 |
- |
- |
PREDICTED: cation/H(+) antiporter 28 [Jatropha curcas] |
| 17 |
Hb_000462_010 |
0.1167826518 |
- |
- |
hypothetical protein VITISV_041696 [Vitis vinifera] |
| 18 |
Hb_009315_010 |
0.1201529509 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 19 |
Hb_001047_210 |
0.1224289349 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 20 |
Hb_001699_110 |
0.1230088117 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |