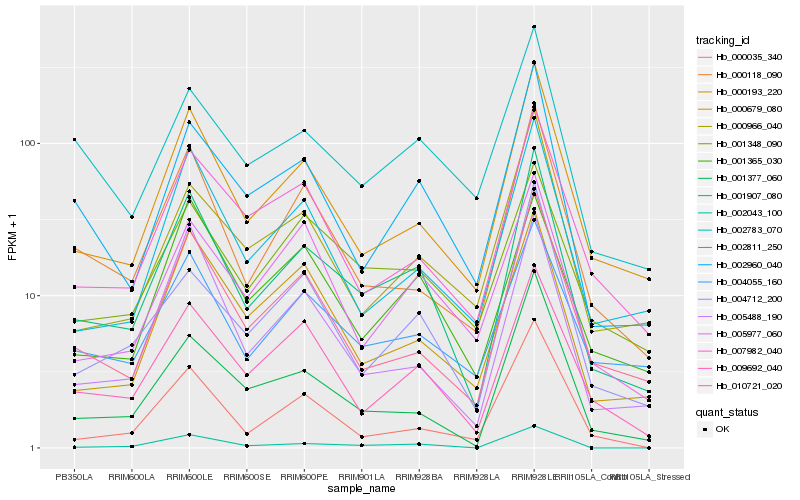
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001907_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628749 [Jatropha curcas] |
| 2 |
Hb_000193_220 |
0.1057271658 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002043_100 |
0.1221089782 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_009692_040 |
0.1233811308 |
- |
- |
PREDICTED: peptide chain release factor PrfB3, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_010721_020 |
0.1239791032 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 4, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002960_040 |
0.1250429435 |
- |
- |
PREDICTED: endoplasmin homolog [Jatropha curcas] |
| 7 |
Hb_000118_090 |
0.1282948149 |
- |
- |
Glutamyl-tRNA reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_001377_060 |
0.1296616865 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01030, mitochondrial [Jatropha curcas] |
| 9 |
Hb_002811_250 |
0.1341389653 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_005488_190 |
0.1369332534 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 11 |
Hb_004712_200 |
0.1389092232 |
- |
- |
hypothetical protein CICLE_v10010292mg [Citrus clementina] |
| 12 |
Hb_004055_160 |
0.142037712 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_007982_040 |
0.1424600487 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000679_080 |
0.1462972888 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000966_040 |
0.1468661177 |
- |
- |
hypothetical protein JCGZ_01642 [Jatropha curcas] |
| 16 |
Hb_002783_070 |
0.1510162096 |
- |
- |
PREDICTED: endoplasmin homolog [Jatropha curcas] |
| 17 |
Hb_001348_090 |
0.1510891551 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 9, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001365_030 |
0.1523777181 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 19 |
Hb_000035_340 |
0.1531745539 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005977_060 |
0.156450724 |
- |
- |
conserved hypothetical protein [Ricinus communis] |