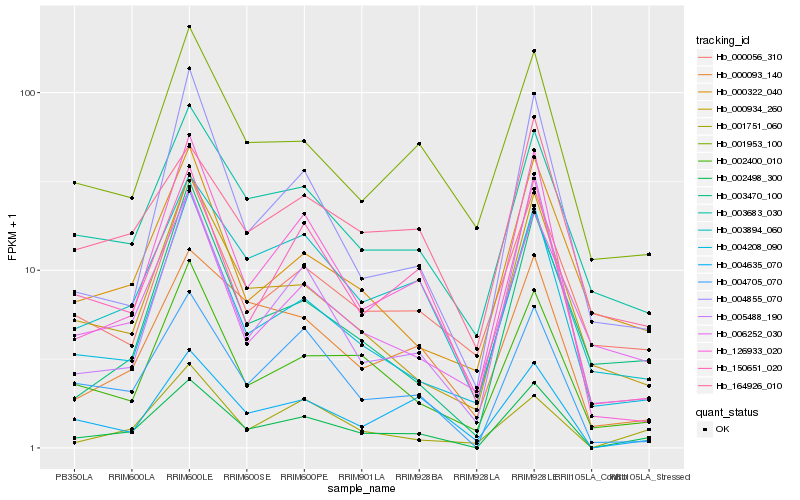
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002498_300 |
0.0 |
- |
- |
BnaC04g50140D [Brassica napus] |
| 2 |
Hb_004705_070 |
0.1467595676 |
- |
- |
PREDICTED: uncharacterized protein LOC105647312 isoform X1 [Jatropha curcas] |
| 3 |
Hb_126933_020 |
0.1488500804 |
- |
- |
PREDICTED: uncharacterized protein LOC105638525 [Jatropha curcas] |
| 4 |
Hb_150651_020 |
0.1555755369 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 5 |
Hb_004208_090 |
0.1563983631 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 6 |
Hb_002400_010 |
0.1643528616 |
- |
- |
PREDICTED: putative L-ascorbate peroxidase 6 [Jatropha curcas] |
| 7 |
Hb_000093_140 |
0.1672039867 |
- |
- |
PREDICTED: fructokinase-like 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_004635_070 |
0.1678498856 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein, chloroplastic [Solanum lycopersicum] |
| 9 |
Hb_005488_190 |
0.1689952621 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 10 |
Hb_003683_030 |
0.1691399962 |
- |
- |
PREDICTED: uridine-cytidine kinase C isoform X1 [Jatropha curcas] |
| 11 |
Hb_000934_260 |
0.1696063733 |
- |
- |
Thylakoid lumenal 29.8 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_003894_060 |
0.17012486 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004855_070 |
0.170268791 |
- |
- |
PLASTID-SPECIFIC RIBOSOMAL protein 4 [Populus trichocarpa] |
| 14 |
Hb_001751_060 |
0.1712895406 |
- |
- |
PREDICTED: SEC12-like protein 1 [Jatropha curcas] |
| 15 |
Hb_000056_310 |
0.176533271 |
- |
- |
PREDICTED: uncharacterized protein LOC105630433 [Jatropha curcas] |
| 16 |
Hb_164926_010 |
0.176837458 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 17 |
Hb_000322_040 |
0.1774376373 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_006252_030 |
0.1776176533 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 19 |
Hb_001953_100 |
0.1776179692 |
- |
- |
PREDICTED: glutamate-1-semialdehyde 2,1-aminomutase 2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_003470_100 |
0.1781152567 |
- |
- |
protein binding protein, putative [Ricinus communis] |