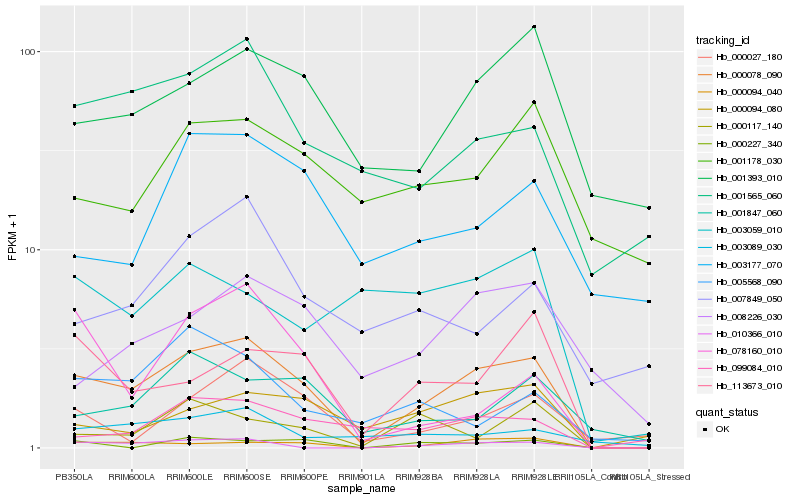
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000078_090 |
0.0 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 2 |
Hb_000094_040 |
0.1820501977 |
- |
- |
- |
| 3 |
Hb_113673_010 |
0.2133815597 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 4 |
Hb_010366_010 |
0.2277290773 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 5 |
Hb_001565_060 |
0.2300049229 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 1, mitochondrial [Jatropha curcas] |
| 6 |
Hb_001393_010 |
0.2362000594 |
- |
- |
hypothetical protein POPTR_0018s06150g [Populus trichocarpa] |
| 7 |
Hb_000094_080 |
0.2471801004 |
- |
- |
PREDICTED: uncharacterized protein LOC105633792 [Jatropha curcas] |
| 8 |
Hb_003089_030 |
0.2474379996 |
- |
- |
3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase, putative [Ricinus communis] |
| 9 |
Hb_001847_060 |
0.2484708945 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Vitis vinifera] |
| 10 |
Hb_099084_010 |
0.2509273596 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 11 |
Hb_000027_180 |
0.2517743118 |
- |
- |
PREDICTED: uncharacterized protein LOC105642058 [Jatropha curcas] |
| 12 |
Hb_008226_030 |
0.2549374328 |
- |
- |
RING-H2 finger protein ATL3C, putative [Ricinus communis] |
| 13 |
Hb_000117_140 |
0.2577912068 |
- |
- |
hypothetical protein CISIN_1g042884mg, partial [Citrus sinensis] |
| 14 |
Hb_003177_070 |
0.259268486 |
- |
- |
PREDICTED: ATP sulfurylase 2 [Jatropha curcas] |
| 15 |
Hb_078160_010 |
0.2612853363 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 16 |
Hb_000227_340 |
0.2619195702 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 17 |
Hb_007849_050 |
0.2636196408 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 18 |
Hb_005568_090 |
0.2641977641 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 19 |
Hb_003059_010 |
0.2646003178 |
- |
- |
PREDICTED: uncharacterized protein LOC104236538 [Nicotiana sylvestris] |
| 20 |
Hb_001178_030 |
0.2656291548 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 2-2, chloroplastic/amyloplastic [Jatropha curcas] |