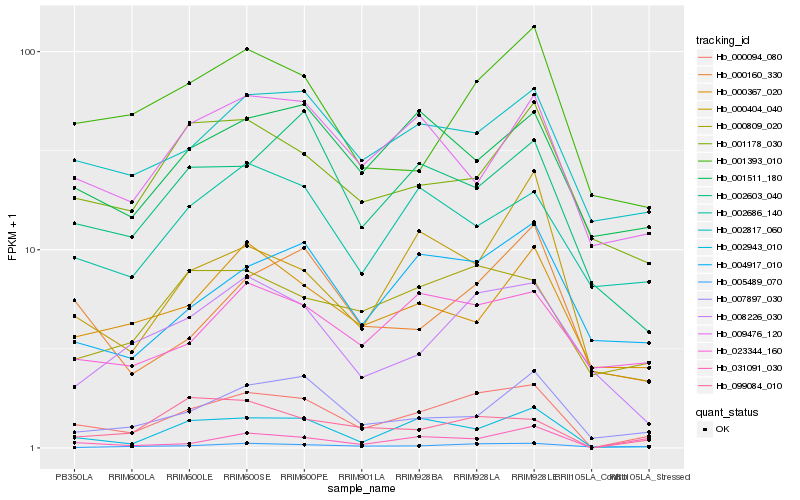
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000094_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633792 [Jatropha curcas] |
| 2 |
Hb_002943_010 |
0.164219162 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Nelumbo nucifera] |
| 3 |
Hb_002817_060 |
0.1734958308 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 4 |
Hb_000809_020 |
0.1795615046 |
desease resistance |
Gene Name: RPW8 |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_001393_010 |
0.1808943808 |
- |
- |
hypothetical protein POPTR_0018s06150g [Populus trichocarpa] |
| 6 |
Hb_004917_010 |
0.1817321309 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Jatropha curcas] |
| 7 |
Hb_007897_030 |
0.1831834967 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR2 [Jatropha curcas] |
| 8 |
Hb_005489_070 |
0.1886280735 |
- |
- |
PREDICTED: transposon Tf2-1 polyprotein isoform X1 [Zea mays] |
| 9 |
Hb_002603_040 |
0.1899518534 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 2-like [Jatropha curcas] |
| 10 |
Hb_000160_330 |
0.1904225749 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 11 |
Hb_099084_010 |
0.1905703231 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 12 |
Hb_008226_030 |
0.1911352311 |
- |
- |
RING-H2 finger protein ATL3C, putative [Ricinus communis] |
| 13 |
Hb_001511_180 |
0.1918272124 |
- |
- |
Clathrin heavy chain 2 -like protein [Gossypium arboreum] |
| 14 |
Hb_001178_030 |
0.1919858855 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 2-2, chloroplastic/amyloplastic [Jatropha curcas] |
| 15 |
Hb_023344_160 |
0.192346683 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 16 |
Hb_000404_040 |
0.1927612726 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_031091_030 |
0.1929696695 |
- |
- |
hypothetical protein OsI_25921 [Oryza sativa Indica Group] |
| 18 |
Hb_002686_140 |
0.1933830991 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000367_020 |
0.1941242704 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 20 |
Hb_009476_120 |
0.1942163446 |
- |
- |
Clathrin heavy chain 1 [Glycine soja] |