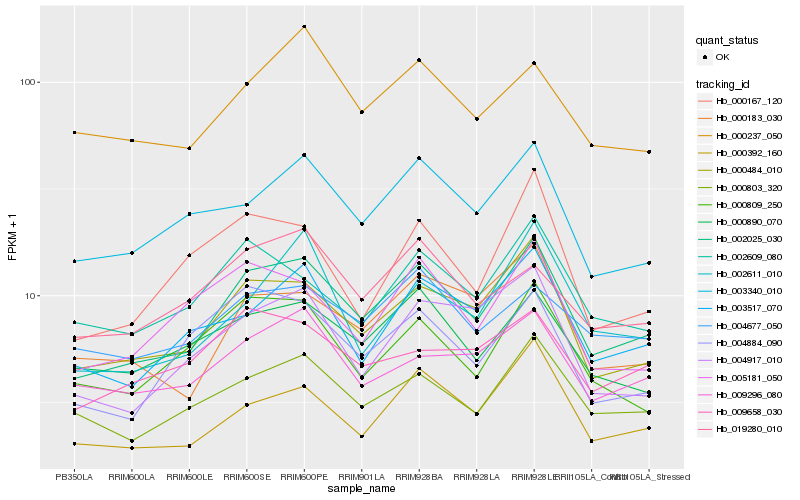
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002025_030 |
0.0 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 2 |
Hb_000484_010 |
0.0688694756 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 23 [Jatropha curcas] |
| 3 |
Hb_000392_160 |
0.0865849285 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000803_320 |
0.0889333437 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 5 |
Hb_003340_010 |
0.091629971 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1 [Vitis vinifera] |
| 6 |
Hb_003517_070 |
0.0949859277 |
- |
- |
PREDICTED: protein transport protein Sec24-like At3g07100 [Jatropha curcas] |
| 7 |
Hb_000183_030 |
0.0968858137 |
- |
- |
PREDICTED: probable zinc transporter protein DDB_G0291141 [Populus euphratica] |
| 8 |
Hb_000890_070 |
0.0998699151 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 9 |
Hb_005181_050 |
0.1037197572 |
- |
- |
PREDICTED: phytochrome B isoform X1 [Jatropha curcas] |
| 10 |
Hb_004917_010 |
0.104051578 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Jatropha curcas] |
| 11 |
Hb_002609_080 |
0.104840106 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_000809_250 |
0.1066290812 |
- |
- |
PREDICTED: uncharacterized protein LOC105639683 [Jatropha curcas] |
| 13 |
Hb_004884_090 |
0.1066977295 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 14 |
Hb_019280_010 |
0.1070536336 |
- |
- |
PREDICTED: F-box protein At5g39450 isoform X2 [Jatropha curcas] |
| 15 |
Hb_004677_050 |
0.1076914089 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_009296_080 |
0.1080774754 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002611_010 |
0.1081003996 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 18 |
Hb_000237_050 |
0.1089101787 |
- |
- |
CP2 [Hevea brasiliensis] |
| 19 |
Hb_009658_030 |
0.1112900232 |
- |
- |
PREDICTED: AP-5 complex subunit beta-1 [Jatropha curcas] |
| 20 |
Hb_000167_120 |
0.1116563129 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |