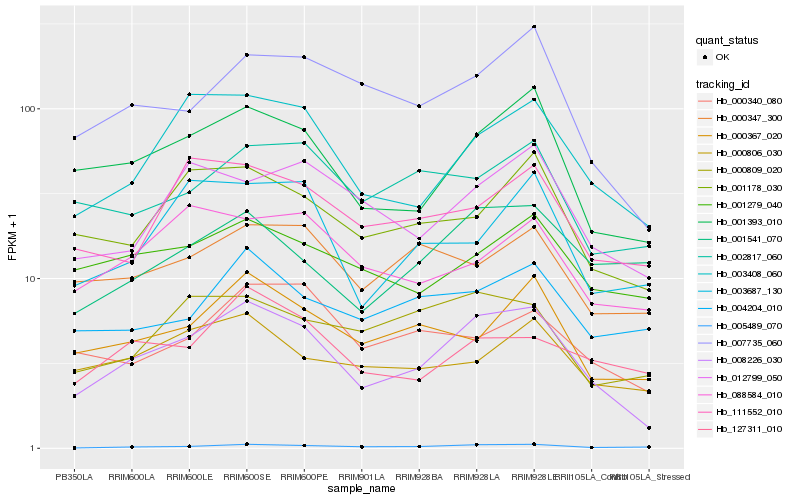
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008226_030 |
0.0 |
- |
- |
RING-H2 finger protein ATL3C, putative [Ricinus communis] |
| 2 |
Hb_001393_010 |
0.1216408763 |
- |
- |
hypothetical protein POPTR_0018s06150g [Populus trichocarpa] |
| 3 |
Hb_003408_060 |
0.1328859195 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 4 |
Hb_005489_070 |
0.1430422539 |
- |
- |
PREDICTED: transposon Tf2-1 polyprotein isoform X1 [Zea mays] |
| 5 |
Hb_007735_060 |
0.1470998216 |
- |
- |
PPase [Hevea brasiliensis] |
| 6 |
Hb_001178_030 |
0.1534655336 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 2-2, chloroplastic/amyloplastic [Jatropha curcas] |
| 7 |
Hb_000367_020 |
0.1589961752 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 8 |
Hb_012799_050 |
0.1657885526 |
- |
- |
PREDICTED: remorin [Jatropha curcas] |
| 9 |
Hb_111552_010 |
0.1677845714 |
- |
- |
PREDICTED: ATP sulfurylase 2 [Jatropha curcas] |
| 10 |
Hb_003687_130 |
0.1683170618 |
- |
- |
PREDICTED: probable methyltransferase PMT3 [Jatropha curcas] |
| 11 |
Hb_000809_020 |
0.1706583663 |
desease resistance |
Gene Name: RPW8 |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_000806_030 |
0.1709924554 |
transcription factor |
TF Family: C2H2 |
PREDICTED: probable lysine-specific demethylase ELF6 [Jatropha curcas] |
| 13 |
Hb_001279_040 |
0.1710022537 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 14 |
Hb_127311_010 |
0.1714804774 |
- |
- |
hypothetical protein JCGZ_07497 [Jatropha curcas] |
| 15 |
Hb_002817_060 |
0.1725648143 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 16 |
Hb_000340_080 |
0.1742011746 |
- |
- |
PREDICTED: isoamylase 2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001541_070 |
0.1743041179 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1-like [Jatropha curcas] |
| 18 |
Hb_000347_300 |
0.1745059161 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 19 |
Hb_088584_010 |
0.1753569175 |
- |
- |
hypothetical protein POPTR_0016s08470g [Populus trichocarpa] |
| 20 |
Hb_004204_010 |
0.1756951918 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |