| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
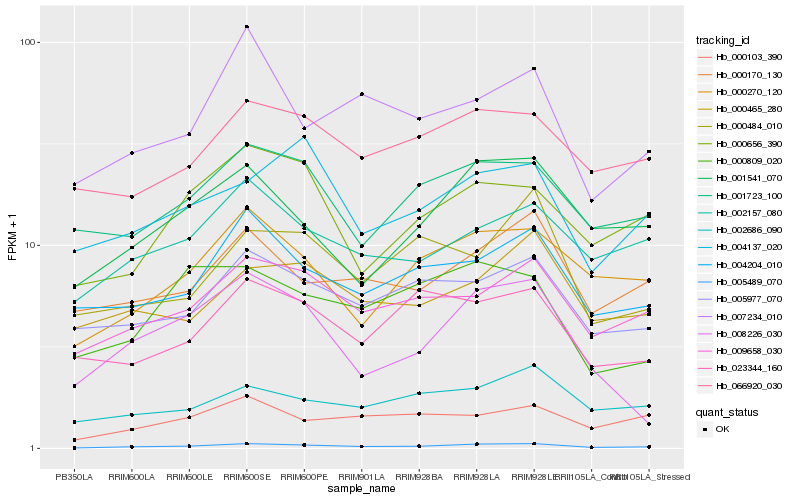
Hb_005489_070 |
0.0 |
- |
- |
PREDICTED: transposon Tf2-1 polyprotein isoform X1 [Zea mays] |
| 2 |
Hb_009658_030 |
0.1168905456 |
- |
- |
PREDICTED: AP-5 complex subunit beta-1 [Jatropha curcas] |
| 3 |
Hb_000270_120 |
0.1224604012 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 4 |
Hb_004204_010 |
0.1288752589 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 5 |
Hb_001541_070 |
0.1299413161 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1-like [Jatropha curcas] |
| 6 |
Hb_005977_070 |
0.1307642044 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000170_130 |
0.1328371618 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 8 |
Hb_001723_100 |
0.1348901257 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002157_080 |
0.1349561183 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 10 |
Hb_066920_030 |
0.1352583208 |
- |
- |
PREDICTED: adagio protein 1 [Jatropha curcas] |
| 11 |
Hb_002686_090 |
0.1360423545 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g13650 [Jatropha curcas] |
| 12 |
Hb_023344_160 |
0.1376734148 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 13 |
Hb_000656_390 |
0.138462426 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000809_020 |
0.1413903075 |
desease resistance |
Gene Name: RPW8 |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_000103_390 |
0.1417652726 |
- |
- |
Pentatricopeptide repeat superfamily protein [Theobroma cacao] |
| 16 |
Hb_000465_280 |
0.1426635741 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g47360 [Jatropha curcas] |
| 17 |
Hb_007234_010 |
0.1427360919 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 18 |
Hb_008226_030 |
0.1430422539 |
- |
- |
RING-H2 finger protein ATL3C, putative [Ricinus communis] |
| 19 |
Hb_000484_010 |
0.1454892149 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 23 [Jatropha curcas] |
| 20 |
Hb_004137_020 |
0.1457267041 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 3 isoform X2 [Jatropha curcas] |