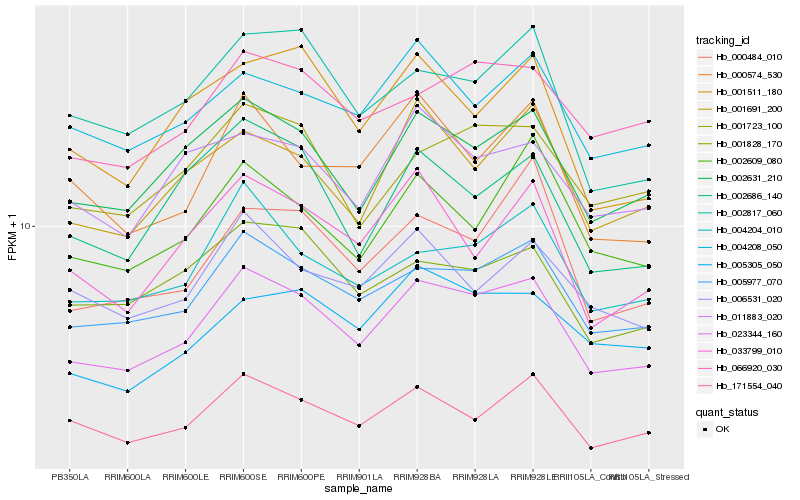
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023344_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 2 |
Hb_005977_070 |
0.0588526129 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002631_210 |
0.0671502026 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 4 |
Hb_004204_010 |
0.0838613732 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 5 |
Hb_001723_100 |
0.0871182361 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001691_200 |
0.0874383583 |
- |
- |
DNA-directed RNA polymerase II subunit RPB2 [Auxenochlorella protothecoides] |
| 7 |
Hb_171554_040 |
0.0891090882 |
- |
- |
PREDICTED: phytanoyl-CoA dioxygenase [Populus euphratica] |
| 8 |
Hb_002817_060 |
0.0897858185 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 9 |
Hb_066920_030 |
0.0904186801 |
- |
- |
PREDICTED: adagio protein 1 [Jatropha curcas] |
| 10 |
Hb_002686_140 |
0.0922070284 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001828_170 |
0.0927826525 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 12 |
Hb_004208_050 |
0.0935641439 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 13 |
Hb_000574_530 |
0.0938898083 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 14 |
Hb_033799_010 |
0.0939296456 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 15 |
Hb_011883_020 |
0.0948512556 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 16 |
Hb_005305_050 |
0.0948949774 |
- |
- |
PREDICTED: armadillo repeat-containing protein 8 [Jatropha curcas] |
| 17 |
Hb_001511_180 |
0.0950548867 |
- |
- |
Clathrin heavy chain 2 -like protein [Gossypium arboreum] |
| 18 |
Hb_002609_080 |
0.0956655616 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_006531_020 |
0.0960276941 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 20 |
Hb_000484_010 |
0.0969896745 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 23 [Jatropha curcas] |