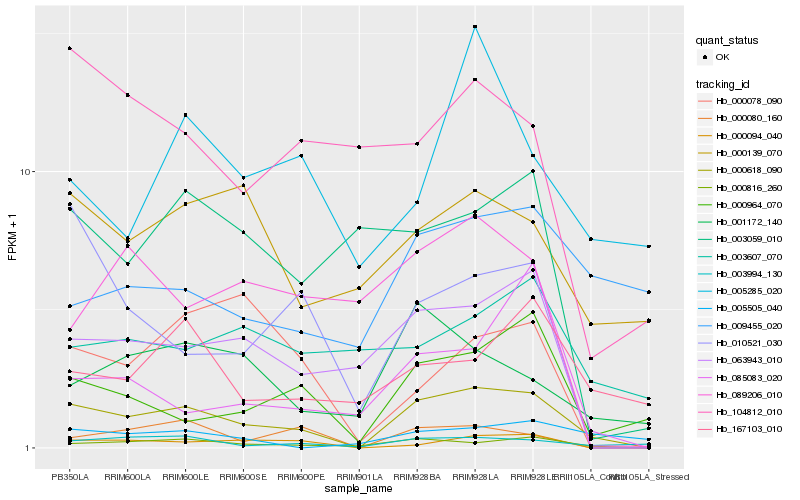
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000618_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_000816_260 |
0.2036678829 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 3 |
Hb_000094_040 |
0.2342242307 |
- |
- |
- |
| 4 |
Hb_003994_130 |
0.2381659002 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1-like [Jatropha curcas] |
| 5 |
Hb_063943_010 |
0.2389453601 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 6 |
Hb_000964_070 |
0.2475758674 |
- |
- |
PREDICTED: ATP synthase subunit O, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000080_160 |
0.255834581 |
- |
- |
Peroxin 13 [Theobroma cacao] |
| 8 |
Hb_001172_140 |
0.2626318164 |
- |
- |
hypothetical protein POPTR_0014s17790g [Populus trichocarpa] |
| 9 |
Hb_000139_070 |
0.2638050466 |
- |
- |
PREDICTED: probable linoleate 9S-lipoxygenase 5 [Jatropha curcas] |
| 10 |
Hb_010521_030 |
0.2661295788 |
- |
- |
- |
| 11 |
Hb_167103_010 |
0.2722448931 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 12 |
Hb_085083_020 |
0.2777319487 |
- |
- |
PREDICTED: receptor-like protein kinase THESEUS 1 [Jatropha curcas] |
| 13 |
Hb_003059_010 |
0.279607113 |
- |
- |
PREDICTED: uncharacterized protein LOC104236538 [Nicotiana sylvestris] |
| 14 |
Hb_000078_090 |
0.2809082716 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 15 |
Hb_104812_010 |
0.2847334105 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 9 [Populus euphratica] |
| 16 |
Hb_009455_020 |
0.2866777101 |
- |
- |
PREDICTED: uncharacterized protein LOC105634581 [Jatropha curcas] |
| 17 |
Hb_089206_010 |
0.2873113674 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 18 |
Hb_005285_020 |
0.2880767865 |
- |
- |
- |
| 19 |
Hb_003607_070 |
0.2893036609 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48910 [Jatropha curcas] |
| 20 |
Hb_005505_040 |
0.2900317013 |
- |
- |
- |