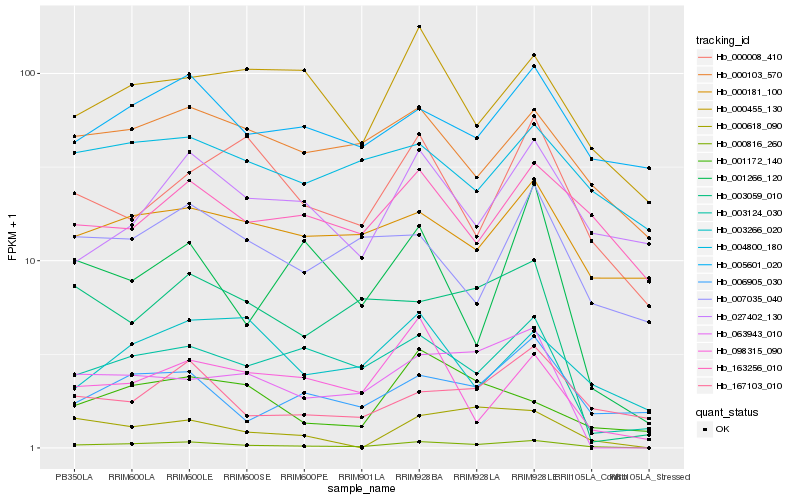
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000816_260 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 2 |
Hb_003124_030 |
0.1803133181 |
- |
- |
At4g33625-like protein [Brassica napus] |
| 3 |
Hb_063943_010 |
0.1896304984 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 4 |
Hb_167103_010 |
0.1965871165 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 5 |
Hb_000103_570 |
0.1977494013 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 6 |
Hb_006905_030 |
0.197948711 |
- |
- |
- |
| 7 |
Hb_000455_130 |
0.2009277563 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_005601_020 |
0.2028709608 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |
| 9 |
Hb_000618_090 |
0.2036678829 |
- |
- |
- |
| 10 |
Hb_163256_010 |
0.2078360154 |
- |
- |
PREDICTED: phosphoglucomutase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000008_410 |
0.2085419791 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 12 |
Hb_007035_040 |
0.2089615888 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003059_010 |
0.2104774594 |
- |
- |
PREDICTED: uncharacterized protein LOC104236538 [Nicotiana sylvestris] |
| 14 |
Hb_098315_090 |
0.2117072368 |
- |
- |
Aldehyde dehydrogenase family 6 member B2 [Morus notabilis] |
| 15 |
Hb_027402_130 |
0.2123413258 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_001172_140 |
0.2128078205 |
- |
- |
hypothetical protein POPTR_0014s17790g [Populus trichocarpa] |
| 17 |
Hb_000181_100 |
0.213056099 |
- |
- |
PREDICTED: protein starmaker [Jatropha curcas] |
| 18 |
Hb_001266_120 |
0.2135422288 |
- |
- |
auxilin, putative [Ricinus communis] |
| 19 |
Hb_004800_180 |
0.2168544629 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 20 |
Hb_003266_020 |
0.2179216623 |
- |
- |
PREDICTED: protein trichome birefringence-like 11 [Jatropha curcas] |