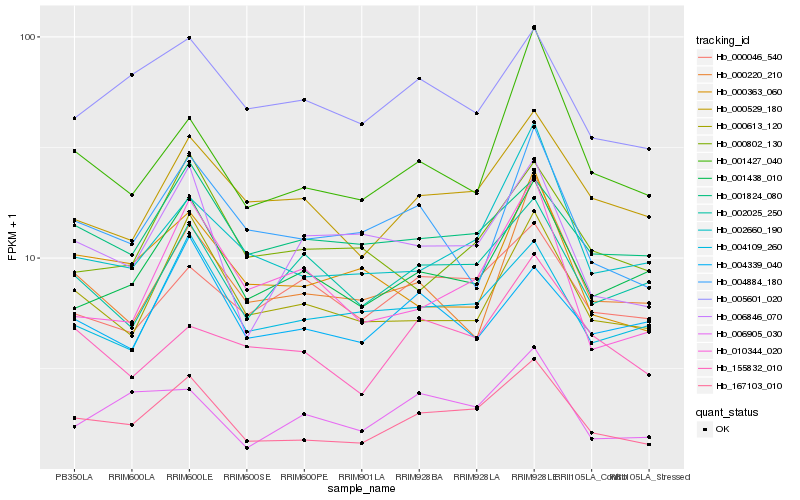
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_167103_010 |
0.0 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 2 |
Hb_000529_180 |
0.117014032 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000613_120 |
0.1183511487 |
transcription factor |
TF Family: SET |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001438_010 |
0.1193591128 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004109_260 |
0.1205627499 |
- |
- |
hypothetical protein JCGZ_06843 [Jatropha curcas] |
| 6 |
Hb_002660_190 |
0.1217048075 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 7 |
Hb_006905_030 |
0.126744234 |
- |
- |
- |
| 8 |
Hb_006846_070 |
0.1271169808 |
- |
- |
PREDICTED: staphylococcal nuclease domain-containing protein 1 [Jatropha curcas] |
| 9 |
Hb_001824_080 |
0.1293178698 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase isoform X1 [Jatropha curcas] |
| 10 |
Hb_005601_020 |
0.1298565016 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |
| 11 |
Hb_001427_040 |
0.1315581103 |
- |
- |
PREDICTED: uncharacterized protein LOC105637784 [Jatropha curcas] |
| 12 |
Hb_002025_250 |
0.1328286957 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 3 [Jatropha curcas] |
| 13 |
Hb_004884_180 |
0.1349147436 |
- |
- |
PREDICTED: protein TIC 56, chloroplastic [Jatropha curcas] |
| 14 |
Hb_155832_010 |
0.1377934946 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 15 |
Hb_010344_020 |
0.1400060959 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g79490, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000802_130 |
0.1408536459 |
- |
- |
hypothetical protein JCGZ_04636 [Jatropha curcas] |
| 17 |
Hb_000363_060 |
0.1410825011 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000220_210 |
0.141247593 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000046_540 |
0.1428144191 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 20 |
Hb_004339_040 |
0.1430012718 |
- |
- |
hypothetical protein POPTR_0162s00290g, partial [Populus trichocarpa] |