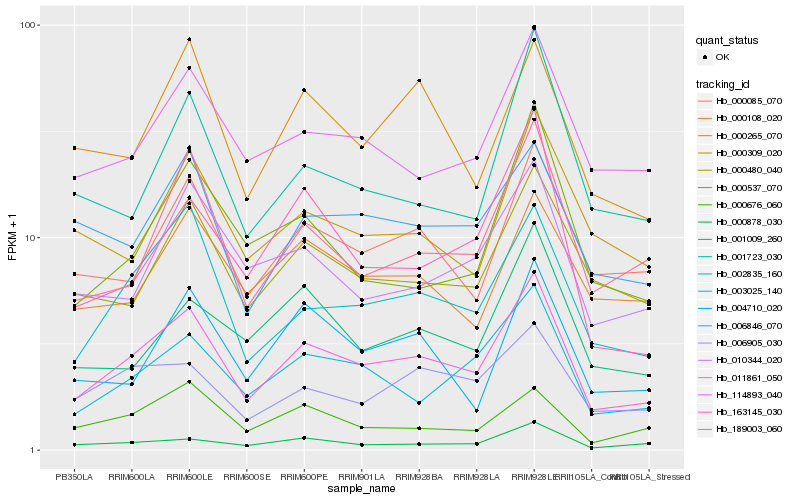
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_114893_040 |
0.0 |
transcription factor |
TF Family: mTERF |
hypothetical protein JCGZ_10155 [Jatropha curcas] |
| 2 |
Hb_002835_160 |
0.1033018811 |
- |
- |
Kinase superfamily protein [Theobroma cacao] |
| 3 |
Hb_003025_140 |
0.1153102854 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000085_070 |
0.1185806528 |
- |
- |
PREDICTED: trigger factor-like protein TIG, Chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_189003_060 |
0.1200567655 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_011861_050 |
0.1233280208 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 7 |
Hb_000480_040 |
0.1244181172 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 8 |
Hb_000878_030 |
0.1254300768 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 9 |
Hb_010344_020 |
0.1308761866 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g79490, mitochondrial [Jatropha curcas] |
| 10 |
Hb_006905_030 |
0.131446721 |
- |
- |
- |
| 11 |
Hb_000265_070 |
0.1322020236 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 12 |
Hb_006846_070 |
0.1331523765 |
- |
- |
PREDICTED: staphylococcal nuclease domain-containing protein 1 [Jatropha curcas] |
| 13 |
Hb_000676_060 |
0.1352637808 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 14 |
Hb_001009_260 |
0.1371148363 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004710_020 |
0.1375260245 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 16 |
Hb_000108_020 |
0.1387911072 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 17 |
Hb_163145_030 |
0.1395018714 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g46580, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000537_070 |
0.140654208 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g25630 [Jatropha curcas] |
| 19 |
Hb_001723_030 |
0.1411511058 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000309_020 |
0.1417213212 |
- |
- |
PREDICTED: uncharacterized protein LOC105641764 [Jatropha curcas] |