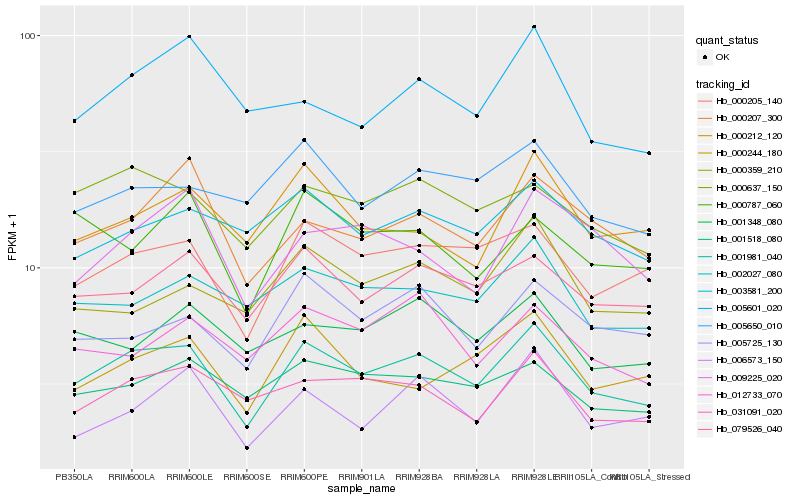
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001981_040 |
0.0 |
transcription factor |
TF Family: PHD |
PREDICTED: origin of replication complex subunit 1B-like [Jatropha curcas] |
| 2 |
Hb_000205_140 |
0.081830903 |
- |
- |
PREDICTED: uncharacterized protein LOC105647751 [Jatropha curcas] |
| 3 |
Hb_079526_040 |
0.0895753283 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 4 |
Hb_000637_150 |
0.0943675311 |
- |
- |
Protein CASP, putative [Ricinus communis] |
| 5 |
Hb_031091_020 |
0.0962860991 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 6 |
Hb_001518_080 |
0.0965298061 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 7 |
Hb_000207_300 |
0.0972239252 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_002027_080 |
0.0984948612 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 9 |
Hb_005725_130 |
0.1001002448 |
- |
- |
PREDICTED: uncharacterized protein KIAA0930 homolog isoform X2 [Jatropha curcas] |
| 10 |
Hb_005650_010 |
0.1036742312 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_003581_200 |
0.1039193234 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |
| 12 |
Hb_000244_180 |
0.1043229072 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000212_120 |
0.1043964166 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 14 |
Hb_006573_150 |
0.1054596569 |
- |
- |
PREDICTED: DNA-directed primase/polymerase protein [Jatropha curcas] |
| 15 |
Hb_001348_080 |
0.1071870257 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 16 |
Hb_012733_070 |
0.1074382207 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 17 |
Hb_009225_020 |
0.1076226198 |
- |
- |
PREDICTED: uncharacterized protein LOC105638232 [Jatropha curcas] |
| 18 |
Hb_000359_210 |
0.1081799852 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005601_020 |
0.1094536739 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |
| 20 |
Hb_000787_060 |
0.1095656341 |
- |
- |
thioredoxin domain-containing protein, putative [Ricinus communis] |