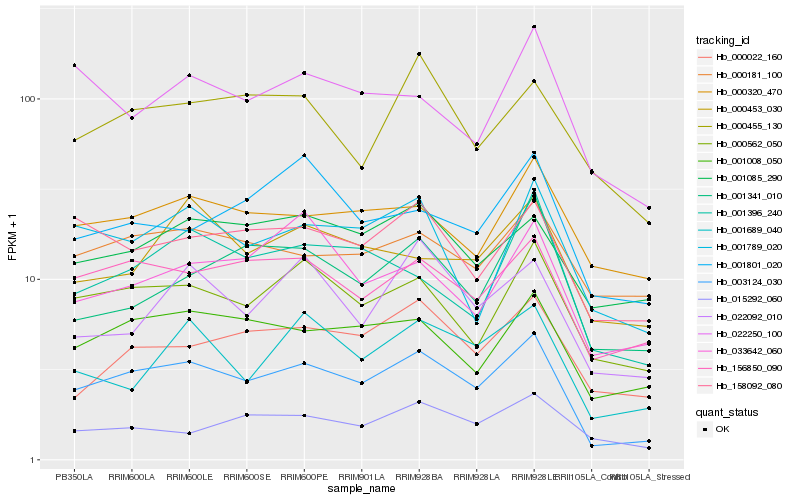
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003124_030 |
0.0 |
- |
- |
At4g33625-like protein [Brassica napus] |
| 2 |
Hb_156850_090 |
0.1141687261 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 3 |
Hb_000562_050 |
0.1174154672 |
- |
- |
PREDICTED: uncharacterized protein LOC105635369 [Jatropha curcas] |
| 4 |
Hb_001008_050 |
0.1203893695 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 5 |
Hb_158092_080 |
0.1308337376 |
- |
- |
hypothetical protein JCGZ_23092 [Jatropha curcas] |
| 6 |
Hb_001789_020 |
0.1333443487 |
- |
- |
PREDICTED: tripeptidyl-peptidase 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001085_290 |
0.1391055554 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 8 |
Hb_000022_160 |
0.1415789771 |
- |
- |
- |
| 9 |
Hb_000320_470 |
0.1427471308 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 10 |
Hb_001689_040 |
0.143321771 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 1-like [Jatropha curcas] |
| 11 |
Hb_001396_240 |
0.1435538789 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_022092_010 |
0.1472038883 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 13 |
Hb_000181_100 |
0.1472688307 |
- |
- |
PREDICTED: protein starmaker [Jatropha curcas] |
| 14 |
Hb_000455_130 |
0.1479958627 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_015292_060 |
0.1483681017 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g38730 [Jatropha curcas] |
| 16 |
Hb_033642_060 |
0.1484883573 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001341_010 |
0.1491842595 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase large chain, chloroplastic [Jatropha curcas] |
| 18 |
Hb_022250_100 |
0.150547973 |
- |
- |
calnexin, putative [Ricinus communis] |
| 19 |
Hb_001801_020 |
0.1506719455 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 20 |
Hb_000453_030 |
0.1509007007 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |