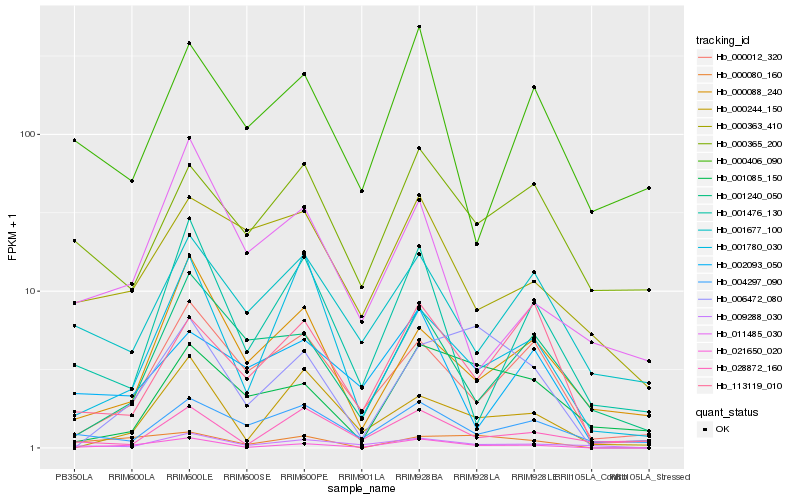
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021650_020 |
0.0 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 2 |
Hb_009288_030 |
0.2324099907 |
- |
- |
PREDICTED: cell division cycle 20.2, cofactor of APC complex-like [Jatropha curcas] |
| 3 |
Hb_000012_320 |
0.2439570232 |
- |
- |
PREDICTED: uncharacterized protein LOC105638164 [Jatropha curcas] |
| 4 |
Hb_000244_150 |
0.2470776339 |
- |
- |
PREDICTED: chromatin assembly factor 1 subunit FAS2 [Jatropha curcas] |
| 5 |
Hb_001476_130 |
0.2492936208 |
- |
- |
PREDICTED: probable protein S-acyltransferase 22 [Jatropha curcas] |
| 6 |
Hb_000080_160 |
0.2520821204 |
- |
- |
Peroxin 13 [Theobroma cacao] |
| 7 |
Hb_006472_080 |
0.254852351 |
- |
- |
- |
| 8 |
Hb_028872_160 |
0.2619098023 |
- |
- |
PREDICTED: ATP-dependent DNA helicase Q-like 5 [Jatropha curcas] |
| 9 |
Hb_004297_090 |
0.2625080904 |
- |
- |
PREDICTED: DNA polymerase epsilon catalytic subunit A [Jatropha curcas] |
| 10 |
Hb_113119_010 |
0.2632990415 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_001677_100 |
0.2652621078 |
- |
- |
delta1-pyrroline-5-carboxylate synthase [Manihot esculenta] |
| 12 |
Hb_001240_050 |
0.2665426447 |
- |
- |
PREDICTED: CSC1-like protein At1g32090 [Jatropha curcas] |
| 13 |
Hb_002093_050 |
0.2666738608 |
- |
- |
hypothetical protein POPTR_0008s02940g [Populus trichocarpa] |
| 14 |
Hb_000406_090 |
0.2693114797 |
- |
- |
unknown [Populus trichocarpa] |
| 15 |
Hb_001780_030 |
0.2696437956 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 16 |
Hb_000088_240 |
0.2707341944 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 17 |
Hb_000363_410 |
0.2709220037 |
- |
- |
hypothetical protein JCGZ_00955 [Jatropha curcas] |
| 18 |
Hb_000365_200 |
0.2727353359 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 19 |
Hb_001085_150 |
0.2738284308 |
- |
- |
Purple acid phosphatase 15 [Theobroma cacao] |
| 20 |
Hb_011485_030 |
0.2751915652 |
- |
- |
PREDICTED: uncharacterized protein LOC105647814 [Jatropha curcas] |