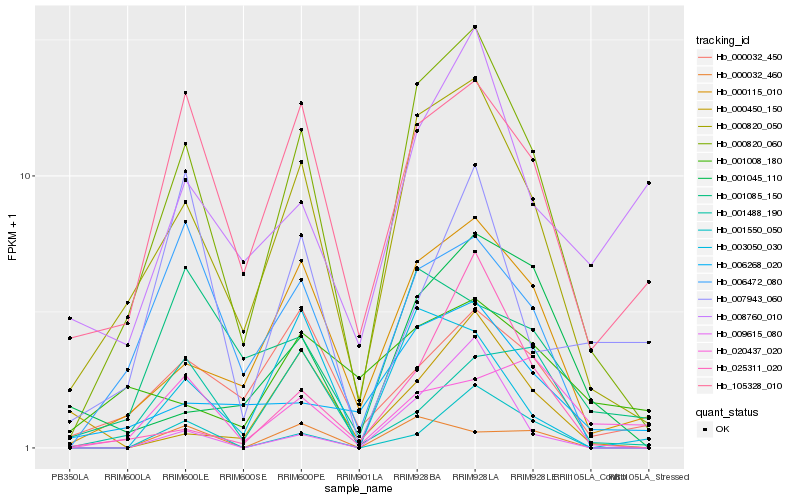
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000820_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_000820_050 |
0.087460842 |
- |
- |
hypothetical protein EUGRSUZ_H02626 [Eucalyptus grandis] |
| 3 |
Hb_000115_010 |
0.1714125584 |
- |
- |
5-methyltetrahydrofolate:homocysteine methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_006472_080 |
0.2154270829 |
- |
- |
- |
| 5 |
Hb_105328_010 |
0.2156465891 |
- |
- |
PREDICTED: low-temperature-induced cysteine proteinase-like [Jatropha curcas] |
| 6 |
Hb_000032_450 |
0.2390684341 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691 [Jatropha curcas] |
| 7 |
Hb_006268_020 |
0.239481511 |
- |
- |
hypothetical protein JCGZ_10516 [Jatropha curcas] |
| 8 |
Hb_001550_050 |
0.2593192419 |
- |
- |
PREDICTED: J domain-containing protein spf31 [Eucalyptus grandis] |
| 9 |
Hb_025311_020 |
0.2602355591 |
- |
- |
PREDICTED: uncharacterized protein LOC105039527 [Elaeis guineensis] |
| 10 |
Hb_001085_150 |
0.2608713725 |
- |
- |
Purple acid phosphatase 15 [Theobroma cacao] |
| 11 |
Hb_001045_110 |
0.263340791 |
transcription factor |
TF Family: M-type |
flowering locus C-like protein [Juglans regia] |
| 12 |
Hb_003050_030 |
0.2686989477 |
- |
- |
Calcyclin-binding protein, putative [Ricinus communis] |
| 13 |
Hb_001488_190 |
0.2763861664 |
- |
- |
alpha-amylase, putative [Ricinus communis] |
| 14 |
Hb_008760_010 |
0.2798418443 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 15 |
Hb_000032_460 |
0.2853102757 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 16 |
Hb_020437_020 |
0.2866855386 |
- |
- |
hypothetical protein POPTR_0001s01410g, partial [Populus trichocarpa] |
| 17 |
Hb_000450_150 |
0.292133393 |
- |
- |
Anthocyanin 5-aromatic acyltransferase, putative [Ricinus communis] |
| 18 |
Hb_001008_180 |
0.2951349338 |
- |
- |
- |
| 19 |
Hb_009615_080 |
0.2990037389 |
- |
- |
hypothetical protein JCGZ_24461 [Jatropha curcas] |
| 20 |
Hb_007943_060 |
0.3060560243 |
- |
- |
Rop5 [Hevea brasiliensis] |