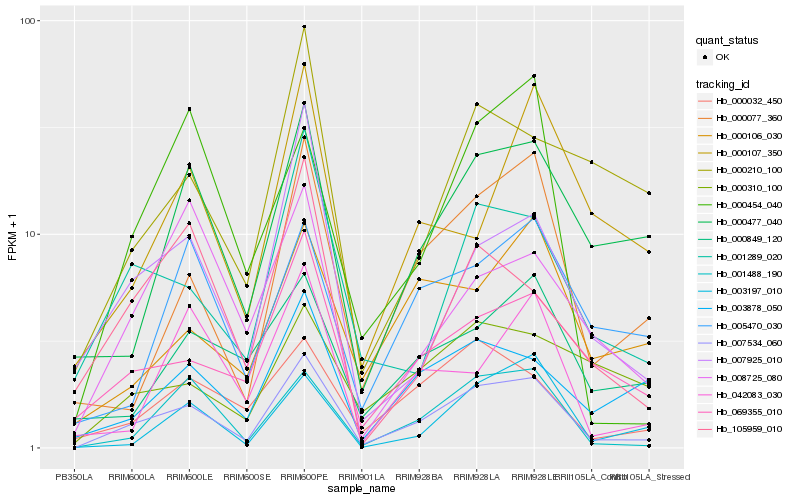
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007534_060 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_001488_190 |
0.170010873 |
- |
- |
alpha-amylase, putative [Ricinus communis] |
| 3 |
Hb_000077_360 |
0.1797201732 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 4 |
Hb_069355_010 |
0.1830127032 |
- |
- |
NBS-LRR resistance protein RGH2 [Manihot esculenta] |
| 5 |
Hb_001289_020 |
0.1918532675 |
- |
- |
PREDICTED: uncharacterized protein LOC105635827 [Jatropha curcas] |
| 6 |
Hb_000477_040 |
0.1972799607 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000032_450 |
0.1998080485 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691 [Jatropha curcas] |
| 8 |
Hb_003878_050 |
0.2000152678 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 9 |
Hb_003197_010 |
0.2153294923 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007925_010 |
0.2165088248 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 1 [Jatropha curcas] |
| 11 |
Hb_005470_030 |
0.2181956438 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000849_120 |
0.2199906557 |
- |
- |
PREDICTED: linoleate 9S-lipoxygenase 6-like [Jatropha curcas] |
| 13 |
Hb_042083_030 |
0.2215400449 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_000106_030 |
0.2226587243 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 15 |
Hb_000454_040 |
0.2292306508 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 16 |
Hb_008725_080 |
0.2300360554 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 7 [Jatropha curcas] |
| 17 |
Hb_000210_100 |
0.2312660904 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 18 |
Hb_105959_010 |
0.2323252835 |
- |
- |
asparagine synthetase, putative [Ricinus communis] |
| 19 |
Hb_000107_350 |
0.2324839197 |
- |
- |
PREDICTED: uncharacterized protein LOC105635576 [Jatropha curcas] |
| 20 |
Hb_000310_100 |
0.2337217087 |
- |
- |
conserved hypothetical protein [Ricinus communis] |