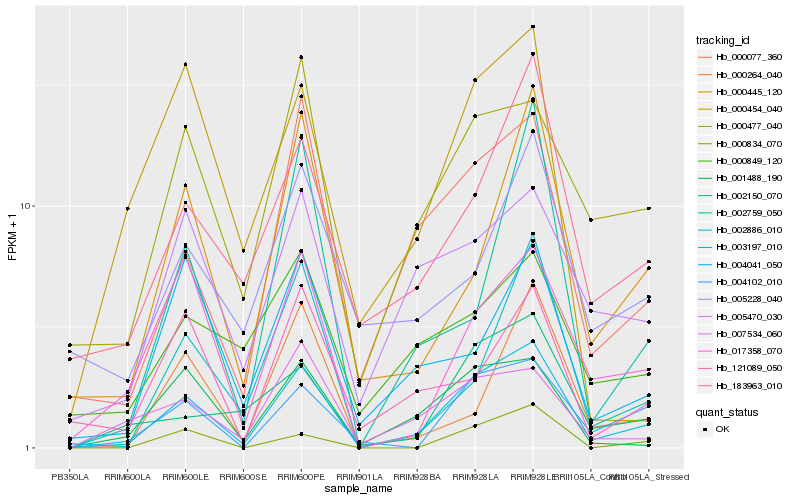
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003197_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000077_360 |
0.1797639426 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 3 |
Hb_004102_010 |
0.1942115707 |
- |
- |
F-box family protein [Theobroma cacao] |
| 4 |
Hb_001488_190 |
0.1981619203 |
- |
- |
alpha-amylase, putative [Ricinus communis] |
| 5 |
Hb_017358_070 |
0.2005795595 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 6 |
Hb_000477_040 |
0.2032485387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000445_120 |
0.205737518 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_183963_010 |
0.2064934746 |
- |
- |
core region of GTP cyclohydrolase I family protein [Populus trichocarpa] |
| 9 |
Hb_007534_060 |
0.2153294923 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_002886_010 |
0.2164074112 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002150_070 |
0.2187511696 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF2-like [Jatropha curcas] |
| 12 |
Hb_005470_030 |
0.2220875813 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000834_070 |
0.2232639172 |
- |
- |
PREDICTED: tankyrase-1 isoform X4 [Jatropha curcas] |
| 14 |
Hb_121089_050 |
0.2261756041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004041_050 |
0.2270054533 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 16 |
Hb_000264_040 |
0.2310371224 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 17 |
Hb_002759_050 |
0.2361670871 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4 [Jatropha curcas] |
| 18 |
Hb_005228_040 |
0.2456842273 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 19 |
Hb_000454_040 |
0.2459364571 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 20 |
Hb_000849_120 |
0.2466983457 |
- |
- |
PREDICTED: linoleate 9S-lipoxygenase 6-like [Jatropha curcas] |