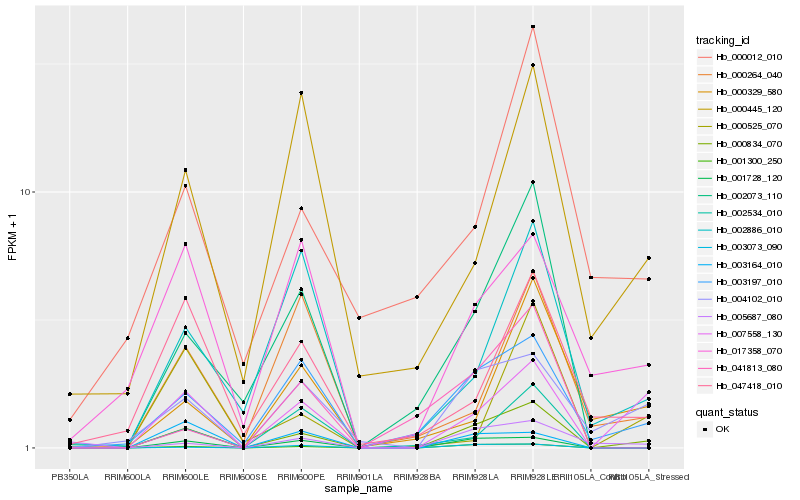
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000834_070 |
0.0 |
- |
- |
PREDICTED: tankyrase-1 isoform X4 [Jatropha curcas] |
| 2 |
Hb_001300_250 |
0.2097935463 |
- |
- |
hypothetical protein JCGZ_22020 [Jatropha curcas] |
| 3 |
Hb_007558_130 |
0.2102025356 |
- |
- |
- |
| 4 |
Hb_003197_010 |
0.2232639172 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001728_120 |
0.2299005902 |
- |
- |
hypothetical protein POPTR_0001s15540g, partial [Populus trichocarpa] |
| 6 |
Hb_004102_010 |
0.2349890195 |
- |
- |
F-box family protein [Theobroma cacao] |
| 7 |
Hb_002073_110 |
0.2715843148 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_047418_010 |
0.2999955646 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein P [Jatropha curcas] |
| 9 |
Hb_002534_010 |
0.3001025448 |
- |
- |
PREDICTED: expansin-A1-like [Jatropha curcas] |
| 10 |
Hb_000329_580 |
0.300719989 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_002886_010 |
0.3019815817 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_017358_070 |
0.3020306248 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 13 |
Hb_005687_080 |
0.3032180652 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 14 |
Hb_003073_090 |
0.3035828549 |
transcription factor |
TF Family: B3 |
hypothetical protein POPTR_0004s23920g [Populus trichocarpa] |
| 15 |
Hb_000445_120 |
0.3136725965 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_000525_070 |
0.3137657 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like [Jatropha curcas] |
| 17 |
Hb_000264_040 |
0.3140597993 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 18 |
Hb_041813_080 |
0.3159347178 |
- |
- |
- |
| 19 |
Hb_000012_010 |
0.3179070374 |
- |
- |
hypothetical protein JCGZ_07709 [Jatropha curcas] |
| 20 |
Hb_003164_010 |
0.3184095757 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |