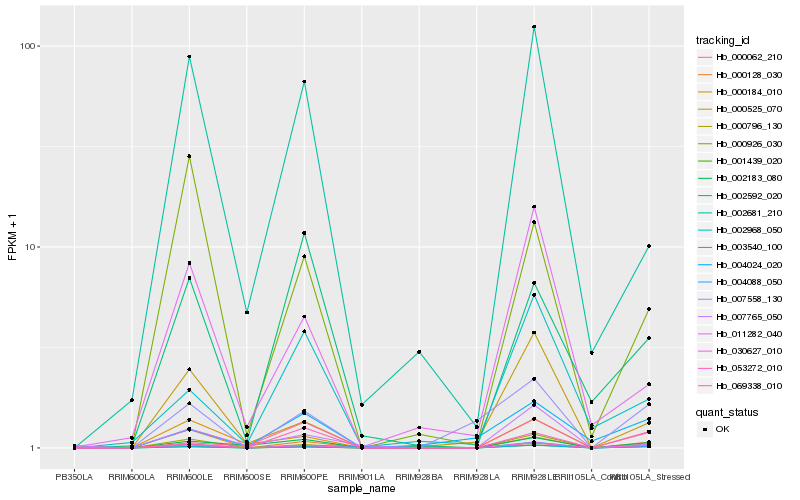
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030627_010 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 [Sesamum indicum] |
| 2 |
Hb_000062_210 |
0.1284295531 |
- |
- |
PREDICTED: BAHD acyltransferase At5g47980-like [Pyrus x bretschneideri] |
| 3 |
Hb_000128_030 |
0.190139834 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000796_130 |
0.2109566604 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 5 |
Hb_002183_080 |
0.2194163365 |
- |
- |
catalytic, putative [Ricinus communis] |
| 6 |
Hb_003540_100 |
0.2349689738 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 7 |
Hb_007558_130 |
0.238086785 |
- |
- |
- |
| 8 |
Hb_000525_070 |
0.2496355067 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like [Jatropha curcas] |
| 9 |
Hb_002681_210 |
0.2565309315 |
- |
- |
PREDICTED: uncharacterized protein LOC105633686 [Jatropha curcas] |
| 10 |
Hb_000184_010 |
0.2566994926 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 11 |
Hb_011282_040 |
0.264999412 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 12 |
Hb_069338_010 |
0.271902125 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000926_030 |
0.274448259 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_001439_020 |
0.2804110348 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 15 |
Hb_002968_050 |
0.2844077245 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 16 |
Hb_053272_010 |
0.2855967084 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 17 |
Hb_004088_050 |
0.289521922 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 18 |
Hb_007765_050 |
0.2895483603 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004024_020 |
0.2903264442 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_002592_020 |
0.291097822 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 isoform X1 [Jatropha curcas] |