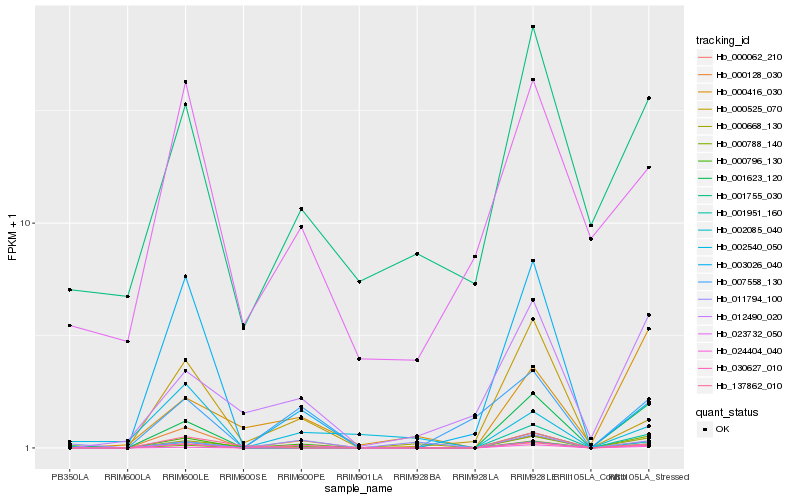
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002085_040 |
0.0 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 2 |
Hb_000796_130 |
0.2382697855 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 3 |
Hb_000062_210 |
0.2432710372 |
- |
- |
PREDICTED: BAHD acyltransferase At5g47980-like [Pyrus x bretschneideri] |
| 4 |
Hb_001623_120 |
0.2684651336 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X2 [Jatropha curcas] |
| 5 |
Hb_030627_010 |
0.3005944516 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 [Sesamum indicum] |
| 6 |
Hb_000668_130 |
0.3122012926 |
- |
- |
- |
| 7 |
Hb_001951_160 |
0.3158702595 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 8 |
Hb_000128_030 |
0.3269293864 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000788_140 |
0.3293907747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_024404_040 |
0.3327907074 |
- |
- |
hypothetical protein JCGZ_22351 [Jatropha curcas] |
| 11 |
Hb_001755_030 |
0.3418523237 |
- |
- |
PREDICTED: D-glycerate 3-kinase, chloroplastic-like [Populus euphratica] |
| 12 |
Hb_023732_050 |
0.350570772 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 13 |
Hb_000416_030 |
0.353536373 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_012490_020 |
0.3591754877 |
- |
- |
PREDICTED: protein phosphatase 2C 77 [Jatropha curcas] |
| 15 |
Hb_137862_010 |
0.3602774032 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 16 |
Hb_002540_050 |
0.3682560417 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g13770, mitochondrial [Jatropha curcas] |
| 17 |
Hb_003026_040 |
0.3697303323 |
- |
- |
Uncharacterized protein TCM_007861 [Theobroma cacao] |
| 18 |
Hb_000525_070 |
0.3701766859 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like [Jatropha curcas] |
| 19 |
Hb_007558_130 |
0.3730859471 |
- |
- |
- |
| 20 |
Hb_011794_100 |
0.3733322837 |
- |
- |
PREDICTED: seven transmembrane domain-containing tyrosine-protein kinase 1-like [Jatropha curcas] |