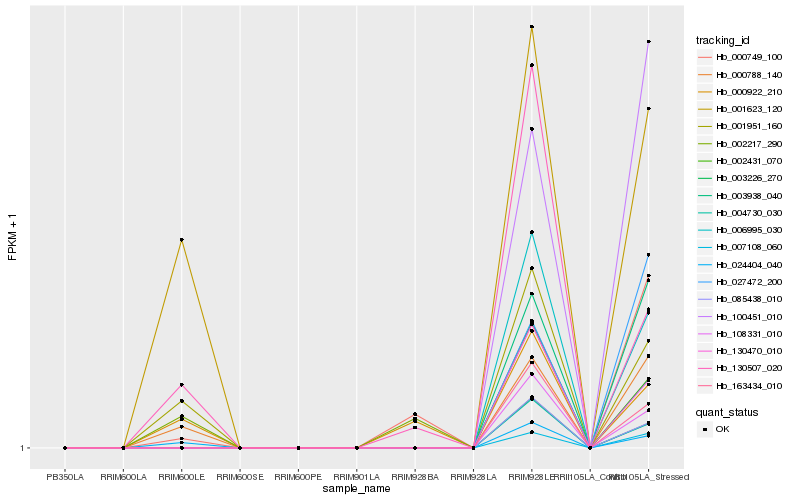
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000788_140 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001951_160 |
0.1219934665 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 3 |
Hb_001623_120 |
0.1270742387 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X2 [Jatropha curcas] |
| 4 |
Hb_024404_040 |
0.1520336824 |
- |
- |
hypothetical protein JCGZ_22351 [Jatropha curcas] |
| 5 |
Hb_003938_040 |
0.2285547192 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF098-like [Jatropha curcas] |
| 6 |
Hb_007108_060 |
0.2285747868 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 7 |
Hb_100451_010 |
0.2357043951 |
- |
- |
- |
| 8 |
Hb_027472_200 |
0.2458373991 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 9 |
Hb_006995_030 |
0.2526956905 |
- |
- |
PREDICTED: uncharacterized protein LOC102663993 [Glycine max] |
| 10 |
Hb_130507_020 |
0.2555934008 |
- |
- |
- |
| 11 |
Hb_000749_100 |
0.2617066488 |
- |
- |
- |
| 12 |
Hb_003226_270 |
0.2635220576 |
- |
- |
hypothetical protein POPTR_0014s12390g [Populus trichocarpa] |
| 13 |
Hb_130470_010 |
0.263740121 |
- |
- |
hypothetical protein, partial [Pseudomonas tolaasii] |
| 14 |
Hb_163434_010 |
0.2683303661 |
- |
- |
hypothetical protein JCGZ_13470 [Jatropha curcas] |
| 15 |
Hb_108331_010 |
0.269643502 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 16 |
Hb_085438_010 |
0.2723064032 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 17 |
Hb_002431_070 |
0.2723552018 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA11 [Jatropha curcas] |
| 18 |
Hb_004730_030 |
0.2724962008 |
- |
- |
synaptonemal complex protein, putative [Ricinus communis] |
| 19 |
Hb_000922_210 |
0.2727976623 |
- |
- |
PREDICTED: 50S ribosomal protein L12, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002217_290 |
0.2730603127 |
- |
- |
hypothetical protein JCGZ_19038 [Jatropha curcas] |