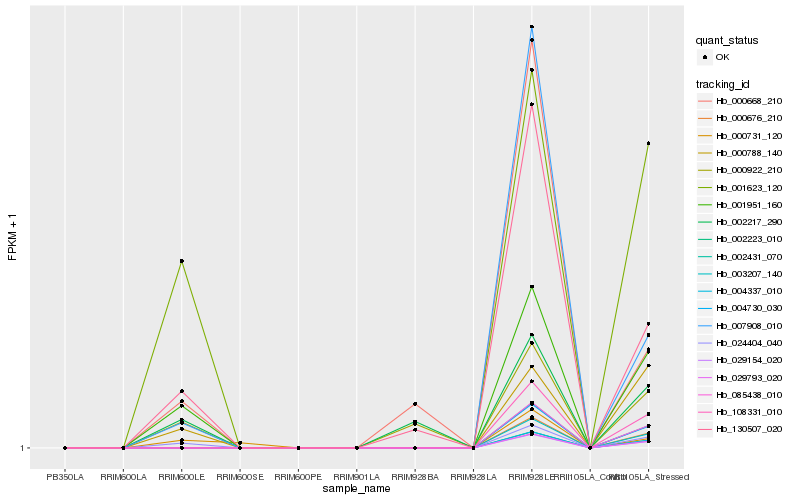
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024404_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_22351 [Jatropha curcas] |
| 2 |
Hb_001951_160 |
0.0362720266 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 3 |
Hb_001623_120 |
0.1221486116 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X2 [Jatropha curcas] |
| 4 |
Hb_130507_020 |
0.148152292 |
- |
- |
- |
| 5 |
Hb_000788_140 |
0.1520336824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007908_010 |
0.1996718786 |
- |
- |
- |
| 7 |
Hb_000731_120 |
0.2383904843 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 8 |
Hb_000922_210 |
0.2395451238 |
- |
- |
PREDICTED: 50S ribosomal protein L12, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002217_290 |
0.2409816744 |
- |
- |
hypothetical protein JCGZ_19038 [Jatropha curcas] |
| 10 |
Hb_000668_210 |
0.2445012818 |
- |
- |
hypothetical protein PHAVU_006G031500g [Phaseolus vulgaris] |
| 11 |
Hb_003207_140 |
0.253776298 |
- |
- |
PREDICTED: protein HOTHEAD [Jatropha curcas] |
| 12 |
Hb_004337_010 |
0.2537765685 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_002223_010 |
0.2537766489 |
- |
- |
hypothetical protein VITISV_009489 [Vitis vinifera] |
| 14 |
Hb_000676_210 |
0.2537779856 |
- |
- |
Cytochrome P450 superfamily protein [Theobroma cacao] |
| 15 |
Hb_029793_020 |
0.2537792135 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 16 |
Hb_029154_020 |
0.2537801008 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_004730_030 |
0.2538140539 |
- |
- |
synaptonemal complex protein, putative [Ricinus communis] |
| 18 |
Hb_002431_070 |
0.2538188416 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA11 [Jatropha curcas] |
| 19 |
Hb_085438_010 |
0.2538205608 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 20 |
Hb_108331_010 |
0.253964131 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |