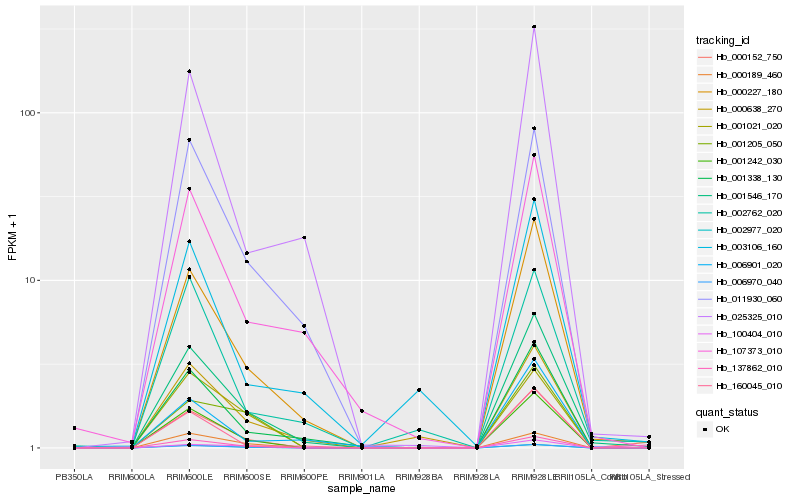
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_750 |
0.0 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 2 |
Hb_006970_040 |
0.1323599254 |
- |
- |
PREDICTED: uncharacterized protein LOC105650489 [Jatropha curcas] |
| 3 |
Hb_137862_010 |
0.1417905897 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_001242_030 |
0.144648888 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000227_180 |
0.1517482721 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001546_170 |
0.154266398 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_002977_020 |
0.179737518 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 1.8-like [Nelumbo nucifera] |
| 8 |
Hb_100404_010 |
0.1861261074 |
- |
- |
Cysteine-rich receptor-like protein kinase 29 [Glycine soja] |
| 9 |
Hb_160045_010 |
0.1914087836 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001338_130 |
0.1925177719 |
- |
- |
hypothetical protein MTR_4g129340 [Medicago truncatula] |
| 11 |
Hb_025325_010 |
0.1981147472 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 12 |
Hb_000189_460 |
0.201411239 |
- |
- |
- |
| 13 |
Hb_011930_060 |
0.20571493 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 14 |
Hb_001205_050 |
0.2081709229 |
- |
- |
- |
| 15 |
Hb_001021_020 |
0.211450683 |
- |
- |
hypothetical protein JCGZ_13899 [Jatropha curcas] |
| 16 |
Hb_002762_020 |
0.2116511288 |
- |
- |
hypothetical protein VITISV_043202 [Vitis vinifera] |
| 17 |
Hb_003106_160 |
0.2125802705 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g67520 [Jatropha curcas] |
| 18 |
Hb_006901_020 |
0.2131529033 |
- |
- |
PREDICTED: guard cell S-type anion channel SLAC1 [Jatropha curcas] |
| 19 |
Hb_000638_270 |
0.2157080617 |
- |
- |
- |
| 20 |
Hb_107373_010 |
0.2167353316 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |