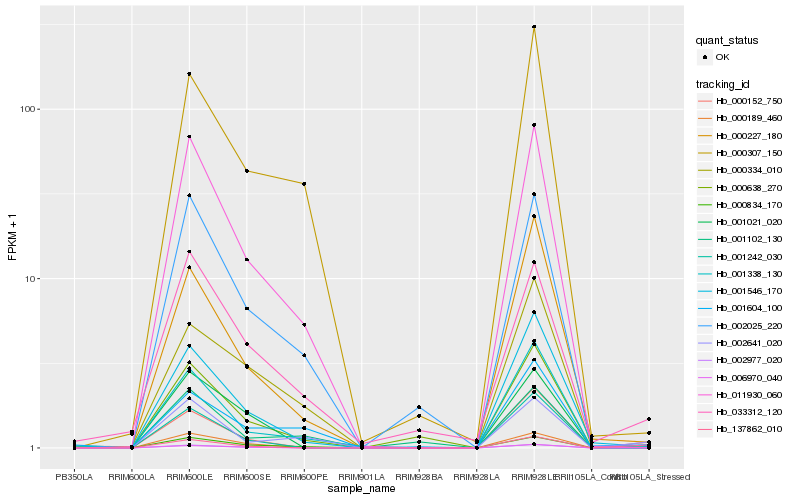
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006970_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650489 [Jatropha curcas] |
| 2 |
Hb_000152_750 |
0.1323599254 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 3 |
Hb_137862_010 |
0.1548067029 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_001546_170 |
0.1678798509 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_011930_060 |
0.1689028068 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 6 |
Hb_002641_020 |
0.1714765765 |
- |
- |
3'-5' exonuclease, putative [Ricinus communis] |
| 7 |
Hb_000227_180 |
0.1759258491 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000334_010 |
0.1769842842 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 9 |
Hb_000834_170 |
0.1777118861 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 10 |
Hb_033312_120 |
0.1797096636 |
- |
- |
PREDICTED: psbQ-like protein 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001604_100 |
0.1857245815 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 12 |
Hb_002977_020 |
0.1858005604 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 1.8-like [Nelumbo nucifera] |
| 13 |
Hb_002025_220 |
0.1869631224 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_000307_150 |
0.1892910741 |
- |
- |
PREDICTED: thioredoxin-like protein CDSP32, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000189_460 |
0.1947925156 |
- |
- |
- |
| 16 |
Hb_001021_020 |
0.1965641898 |
- |
- |
hypothetical protein JCGZ_13899 [Jatropha curcas] |
| 17 |
Hb_000638_270 |
0.1977337371 |
- |
- |
- |
| 18 |
Hb_001338_130 |
0.1980424132 |
- |
- |
hypothetical protein MTR_4g129340 [Medicago truncatula] |
| 19 |
Hb_001242_030 |
0.1990495574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001102_130 |
0.2007684532 |
transcription factor |
TF Family: PHD |
PREDICTED: protein Jade-1 [Populus euphratica] |