| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
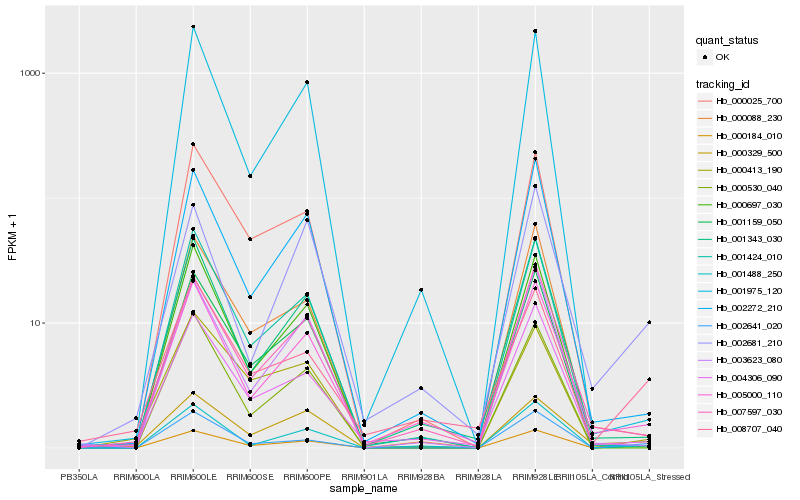
Hb_000184_010 |
0.0 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 2 |
Hb_002641_020 |
0.1057676555 |
- |
- |
3'-5' exonuclease, putative [Ricinus communis] |
| 3 |
Hb_001488_250 |
0.1404107539 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005000_110 |
0.150227277 |
- |
- |
PREDICTED: uncharacterized protein LOC105637872 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002272_210 |
0.1542232036 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000329_500 |
0.1549369777 |
- |
- |
PREDICTED: uncharacterized protein LOC105643180 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001159_050 |
0.1577302817 |
- |
- |
PREDICTED: uncharacterized protein LOC105645952 [Jatropha curcas] |
| 8 |
Hb_008707_040 |
0.1579277241 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000530_040 |
0.159861444 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_007597_030 |
0.1623987161 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_001343_030 |
0.1633699295 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit N, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000413_190 |
0.1638731671 |
- |
- |
PREDICTED: oxygen-evolving enhancer protein 3-2, chloroplastic [Pyrus x bretschneideri] |
| 13 |
Hb_001424_010 |
0.1641436671 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 14 |
Hb_000697_030 |
0.1661818322 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Pyrus x bretschneideri] |
| 15 |
Hb_000088_230 |
0.1667009794 |
- |
- |
hypothetical protein POPTR_0013s11340g [Populus trichocarpa] |
| 16 |
Hb_004306_090 |
0.1673343002 |
- |
- |
PREDICTED: uncharacterized protein LOC105644392 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000025_700 |
0.1676431318 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 18 |
Hb_001975_120 |
0.1684299549 |
- |
- |
chlorophyll a/b binding protein type II [Glycine max] |
| 19 |
Hb_002681_210 |
0.1685008423 |
- |
- |
PREDICTED: uncharacterized protein LOC105633686 [Jatropha curcas] |
| 20 |
Hb_003623_080 |
0.1686277224 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Populus euphratica] |