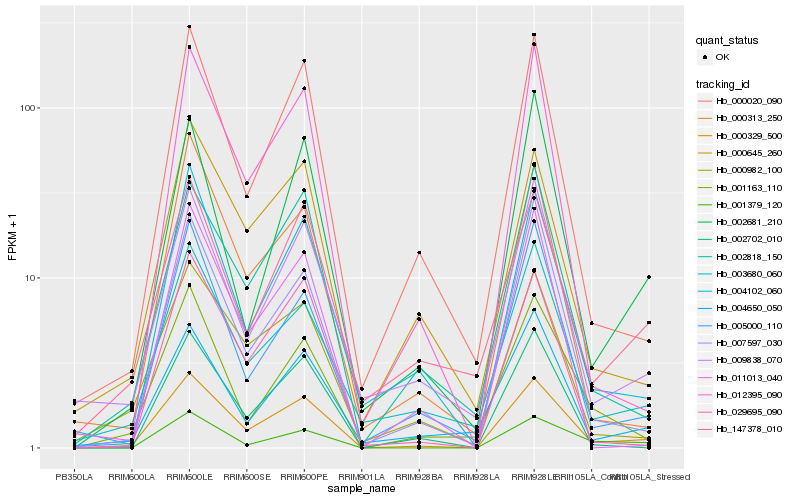
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_500 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643180 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000020_090 |
0.1259905225 |
- |
- |
hypothetical protein JCGZ_09084 [Jatropha curcas] |
| 3 |
Hb_004102_060 |
0.1278659124 |
- |
- |
JHL25H03.11 [Jatropha curcas] |
| 4 |
Hb_003680_060 |
0.1313083002 |
- |
- |
hypothetical protein VITISV_022322 [Vitis vinifera] |
| 5 |
Hb_002818_150 |
0.132952612 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0014s10680g [Populus trichocarpa] |
| 6 |
Hb_000982_100 |
0.1356315263 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor PIF1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_005000_110 |
0.1373396419 |
- |
- |
PREDICTED: uncharacterized protein LOC105637872 isoform X1 [Jatropha curcas] |
| 8 |
Hb_009838_070 |
0.1374108145 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000645_260 |
0.1377142397 |
- |
- |
PREDICTED: uncharacterized protein LOC105643508 [Jatropha curcas] |
| 10 |
Hb_007597_030 |
0.1381823842 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_012395_090 |
0.1386094984 |
- |
- |
PREDICTED: protein PROTON GRADIENT REGULATION 5, chloroplastic [Jatropha curcas] |
| 12 |
Hb_029695_090 |
0.1430906387 |
- |
- |
PREDICTED: probable inactive patatin-like protein 9 [Jatropha curcas] |
| 13 |
Hb_001163_110 |
0.1432746363 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PKL2 [Jatropha curcas] |
| 14 |
Hb_002702_010 |
0.1465800928 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 15 |
Hb_002681_210 |
0.1466601127 |
- |
- |
PREDICTED: uncharacterized protein LOC105633686 [Jatropha curcas] |
| 16 |
Hb_000313_250 |
0.1473595442 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 17 |
Hb_004650_050 |
0.1474836788 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_147378_010 |
0.1480401447 |
- |
- |
- |
| 19 |
Hb_001379_120 |
0.1497836667 |
- |
- |
hypothetical protein POPTR_0001s29730g [Populus trichocarpa] |
| 20 |
Hb_011013_040 |
0.1522104062 |
transcription factor |
TF Family: C2C2-YABBY |
unknown [Populus trichocarpa] |