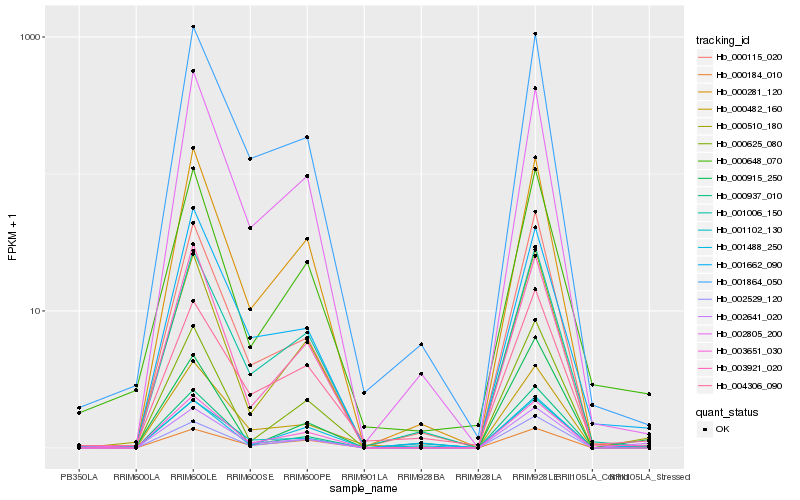
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002641_020 |
0.0 |
- |
- |
3'-5' exonuclease, putative [Ricinus communis] |
| 2 |
Hb_000184_010 |
0.1057676555 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 3 |
Hb_000510_180 |
0.1369915903 |
- |
- |
unknown [Lotus japonicus] |
| 4 |
Hb_001102_130 |
0.1393554939 |
transcription factor |
TF Family: PHD |
PREDICTED: protein Jade-1 [Populus euphratica] |
| 5 |
Hb_001488_250 |
0.143091129 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001662_090 |
0.1445429686 |
- |
- |
PREDICTED: thiosulfate sulfurtransferase 16, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_001864_050 |
0.1478833376 |
- |
- |
chloroplast oxygen-evolving enhancer protein [Manihot esculenta] |
| 8 |
Hb_003921_020 |
0.1500580211 |
- |
- |
hypothetical protein JCGZ_10628 [Jatropha curcas] |
| 9 |
Hb_001006_150 |
0.1503425798 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_000115_020 |
0.1505593175 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger family protein [Populus trichocarpa] |
| 11 |
Hb_000915_250 |
0.1508191763 |
- |
- |
chloroplast latex aldolase-like protein [Manihot esculenta] |
| 12 |
Hb_000937_010 |
0.1509423704 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000625_080 |
0.1510771408 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000281_120 |
0.1523516543 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_000648_070 |
0.1524920799 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 16 |
Hb_003651_030 |
0.1531848847 |
- |
- |
lyase, putative [Ricinus communis] |
| 17 |
Hb_002529_120 |
0.1535816878 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7-like [Jatropha curcas] |
| 18 |
Hb_002805_200 |
0.1539373932 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 19 |
Hb_000482_160 |
0.1546596815 |
- |
- |
hypothetical protein JCGZ_07670 [Jatropha curcas] |
| 20 |
Hb_004306_090 |
0.1548980038 |
- |
- |
PREDICTED: uncharacterized protein LOC105644392 isoform X2 [Jatropha curcas] |