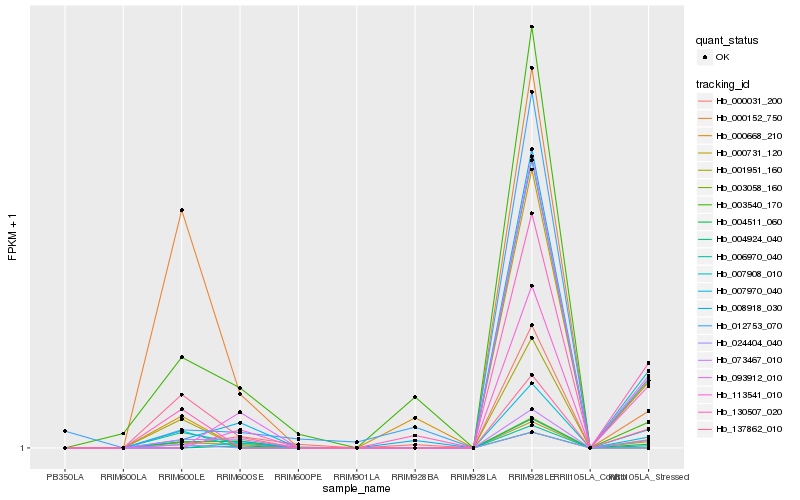
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000731_120 |
0.0 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 2 |
Hb_137862_010 |
0.2200412244 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 3 |
Hb_024404_040 |
0.2383904843 |
- |
- |
hypothetical protein JCGZ_22351 [Jatropha curcas] |
| 4 |
Hb_000152_750 |
0.2407592093 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 5 |
Hb_130507_020 |
0.24776571 |
- |
- |
- |
| 6 |
Hb_001951_160 |
0.2553989011 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 7 |
Hb_004511_060 |
0.2559853736 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103328625 [Prunus mume] |
| 8 |
Hb_007908_010 |
0.2601356093 |
- |
- |
- |
| 9 |
Hb_093912_010 |
0.2603232022 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 10 |
Hb_113541_010 |
0.2666249465 |
- |
- |
PREDICTED: uncharacterized protein LOC105762713 [Gossypium raimondii] |
| 11 |
Hb_007970_040 |
0.2679542506 |
- |
- |
PREDICTED: uncharacterized protein LOC104587672 [Nelumbo nucifera] |
| 12 |
Hb_006970_040 |
0.2717025917 |
- |
- |
PREDICTED: uncharacterized protein LOC105650489 [Jatropha curcas] |
| 13 |
Hb_000031_200 |
0.2812617405 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM3-like [Jatropha curcas] |
| 14 |
Hb_003540_170 |
0.281904515 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21705, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_000668_210 |
0.2857548205 |
- |
- |
hypothetical protein PHAVU_006G031500g [Phaseolus vulgaris] |
| 16 |
Hb_008918_030 |
0.2862812647 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 17 |
Hb_012753_070 |
0.289220259 |
- |
- |
hypothetical protein VITISV_033646 [Vitis vinifera] |
| 18 |
Hb_073467_010 |
0.2906733421 |
- |
- |
PREDICTED: uncharacterized protein LOC105634453 [Jatropha curcas] |
| 19 |
Hb_004924_040 |
0.2913636974 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 20 |
Hb_003058_160 |
0.2914219471 |
- |
- |
conserved hypothetical protein [Ricinus communis] |