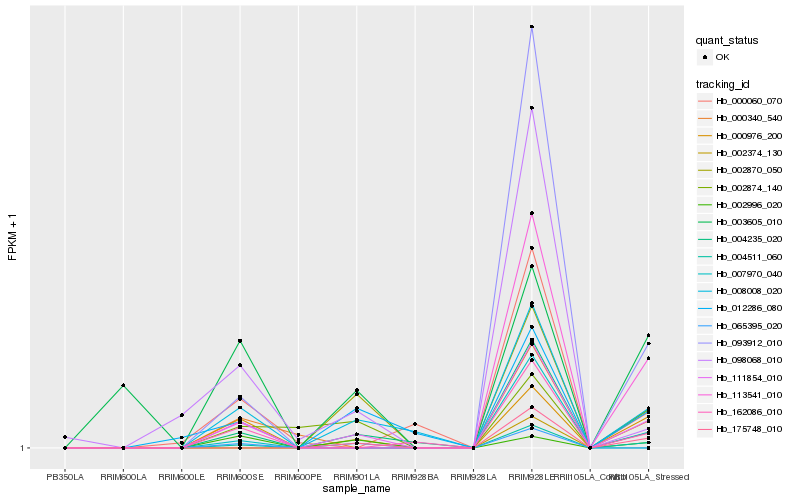
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_111854_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 2 |
Hb_008008_020 |
0.1775079959 |
- |
- |
- |
| 3 |
Hb_004511_060 |
0.2108211952 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103328625 [Prunus mume] |
| 4 |
Hb_093912_010 |
0.2288781226 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 5 |
Hb_113541_010 |
0.2357824857 |
- |
- |
PREDICTED: uncharacterized protein LOC105762713 [Gossypium raimondii] |
| 6 |
Hb_000976_200 |
0.2427329334 |
- |
- |
- |
| 7 |
Hb_007970_040 |
0.2439332844 |
- |
- |
PREDICTED: uncharacterized protein LOC104587672 [Nelumbo nucifera] |
| 8 |
Hb_162086_010 |
0.2520679453 |
- |
- |
PREDICTED: uncharacterized protein LOC105634453 [Jatropha curcas] |
| 9 |
Hb_012286_080 |
0.25704655 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 2 [Jatropha curcas] |
| 10 |
Hb_000340_540 |
0.2729818171 |
- |
- |
PREDICTED: peroxidase 39-like [Jatropha curcas] |
| 11 |
Hb_175748_010 |
0.2743785082 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 12 |
Hb_098068_010 |
0.2748922921 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 8 [Jatropha curcas] |
| 13 |
Hb_002874_140 |
0.2884185469 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 11 homolog [Prunus mume] |
| 14 |
Hb_065395_020 |
0.2986800646 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 15 |
Hb_000060_070 |
0.3006179625 |
- |
- |
hypothetical protein EUTSA_v10023188mg, partial [Eutrema salsugineum] |
| 16 |
Hb_002996_020 |
0.3014407551 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 17 |
Hb_003605_010 |
0.3029116231 |
- |
- |
- |
| 18 |
Hb_002374_130 |
0.3064971494 |
- |
- |
PREDICTED: metal transporter Nramp5-like [Populus euphratica] |
| 19 |
Hb_002870_050 |
0.3130948998 |
- |
- |
- |
| 20 |
Hb_004235_020 |
0.3217669188 |
- |
- |
PREDICTED: uncharacterized protein LOC103322555 [Prunus mume] |