| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
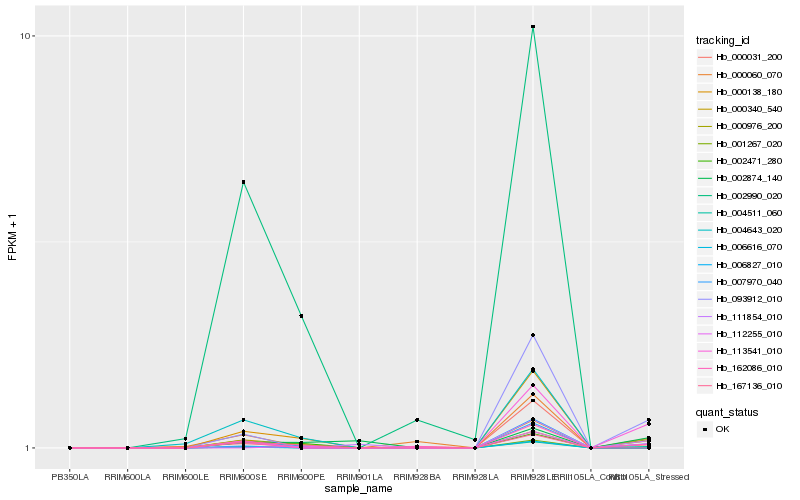
Hb_000340_540 |
0.0 |
- |
- |
PREDICTED: peroxidase 39-like [Jatropha curcas] |
| 2 |
Hb_004511_060 |
0.2188047412 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103328625 [Prunus mume] |
| 3 |
Hb_000976_200 |
0.2304361075 |
- |
- |
- |
| 4 |
Hb_093912_010 |
0.2403406479 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 5 |
Hb_000060_070 |
0.2460207068 |
- |
- |
hypothetical protein EUTSA_v10023188mg, partial [Eutrema salsugineum] |
| 6 |
Hb_113541_010 |
0.2477205791 |
- |
- |
PREDICTED: uncharacterized protein LOC105762713 [Gossypium raimondii] |
| 7 |
Hb_162086_010 |
0.2496158561 |
- |
- |
PREDICTED: uncharacterized protein LOC105634453 [Jatropha curcas] |
| 8 |
Hb_007970_040 |
0.256574254 |
- |
- |
PREDICTED: uncharacterized protein LOC104587672 [Nelumbo nucifera] |
| 9 |
Hb_111854_010 |
0.2729818171 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 10 |
Hb_000031_200 |
0.2774791781 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM3-like [Jatropha curcas] |
| 11 |
Hb_000138_180 |
0.2883170113 |
- |
- |
PREDICTED: uncharacterized protein LOC105131773 [Populus euphratica] |
| 12 |
Hb_006827_010 |
0.2917969497 |
- |
- |
retrotransposon protein, putative, unclassified, expressed [Oryza sativa Japonica Group] |
| 13 |
Hb_001267_020 |
0.2967382591 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 14 |
Hb_112255_010 |
0.2984092294 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 15 |
Hb_167136_010 |
0.2993528019 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 16 |
Hb_002990_020 |
0.2996775886 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 17 |
Hb_006616_070 |
0.3026678855 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_004643_020 |
0.3045207849 |
- |
- |
hypothetical protein AMTR_s05716p00004100, partial [Amborella trichopoda] |
| 19 |
Hb_002471_280 |
0.3097315715 |
- |
- |
PREDICTED: cyclin-SDS [Jatropha curcas] |
| 20 |
Hb_002874_140 |
0.3099445922 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 11 homolog [Prunus mume] |