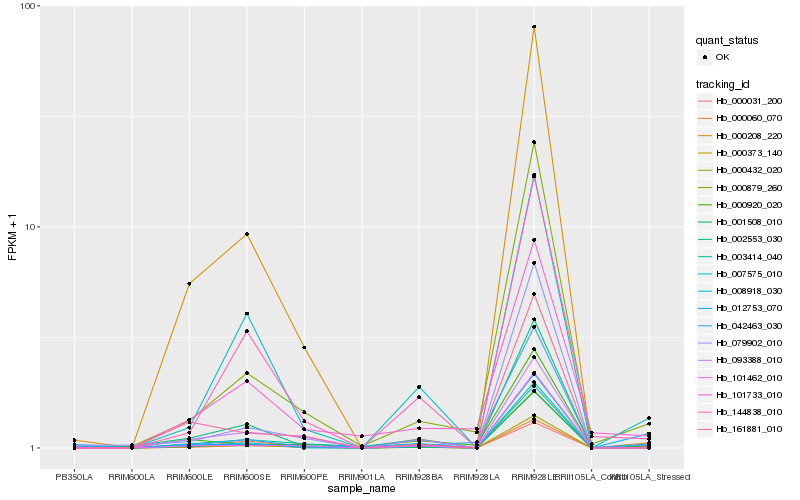
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000031_200 |
0.0 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM3-like [Jatropha curcas] |
| 2 |
Hb_008918_030 |
0.1105176287 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 3 |
Hb_000879_260 |
0.1351511281 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g43190 [Jatropha curcas] |
| 4 |
Hb_007575_010 |
0.1491244288 |
- |
- |
PREDICTED: cytochrome P450 71A1 [Vitis vinifera] |
| 5 |
Hb_144838_010 |
0.164834766 |
- |
- |
hypothetical protein JCGZ_08246 [Jatropha curcas] |
| 6 |
Hb_000373_140 |
0.1861527965 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 7 |
Hb_001508_010 |
0.1863353141 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_042463_030 |
0.1872537645 |
- |
- |
unknown [Lotus japonicus] |
| 9 |
Hb_101462_010 |
0.1881747558 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 10 |
Hb_012753_070 |
0.1884642384 |
- |
- |
hypothetical protein VITISV_033646 [Vitis vinifera] |
| 11 |
Hb_000060_070 |
0.188504151 |
- |
- |
hypothetical protein EUTSA_v10023188mg, partial [Eutrema salsugineum] |
| 12 |
Hb_003414_040 |
0.1903864788 |
- |
- |
hypothetical protein VITISV_016691 [Vitis vinifera] |
| 13 |
Hb_000432_020 |
0.1909572575 |
desease resistance |
Gene Name: Tropomyosin_1 |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000208_220 |
0.1932143038 |
- |
- |
- |
| 15 |
Hb_002553_030 |
0.193539618 |
- |
- |
hypothetical protein POPTR_0008s16550g [Populus trichocarpa] |
| 16 |
Hb_101733_010 |
0.1944367417 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000920_020 |
0.1961979515 |
- |
- |
OSJNBa0004L19.22 [Oryza sativa Japonica Group] |
| 18 |
Hb_093388_010 |
0.1999258818 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 19 |
Hb_079902_010 |
0.2000048353 |
- |
- |
cytochrome c biogenesis B (mitochondrion) [Hevea brasiliensis] |
| 20 |
Hb_161881_010 |
0.2001567454 |
- |
- |
laccase, putative [Ricinus communis] |