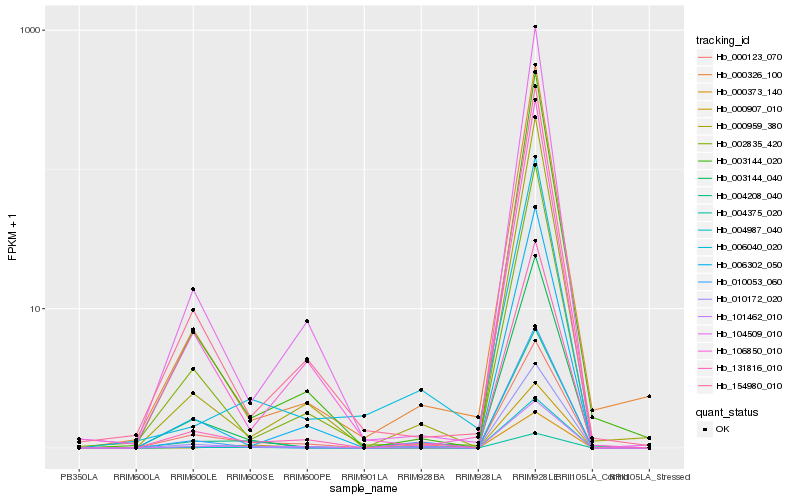
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004208_040 |
0.0 |
- |
- |
UDP-glycosyltransferase 86A1 [Morus notabilis] |
| 2 |
Hb_000373_140 |
0.0702573525 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 3 |
Hb_004987_040 |
0.0761423935 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 4 |
Hb_003144_040 |
0.0800766848 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 5 |
Hb_131816_010 |
0.0834981768 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 6 |
Hb_010172_020 |
0.0895723196 |
- |
- |
PREDICTED: tyrosine/DOPA decarboxylase 1-like [Jatropha curcas] |
| 7 |
Hb_101462_010 |
0.0957154775 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003144_020 |
0.0959415603 |
- |
- |
(R)-limonene synthase, putative [Ricinus communis] |
| 9 |
Hb_004375_020 |
0.0990009256 |
- |
- |
hypothetical protein VITISV_034560 [Vitis vinifera] |
| 10 |
Hb_010053_060 |
0.1010960248 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 11 |
Hb_000123_070 |
0.1012487923 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 12 |
Hb_000326_100 |
0.1023260136 |
transcription factor |
TF Family: MYB-related |
DnaJ homolog subfamily C member 2 [Morus notabilis] |
| 13 |
Hb_154980_010 |
0.1037965591 |
- |
- |
photosystem I P700 chlorophyll A apoprotein A2 [Medicago truncatula] |
| 14 |
Hb_002835_420 |
0.1060029565 |
- |
- |
photosystem I P700 chlorophyll A apoprotein A2 [Medicago truncatula] |
| 15 |
Hb_000959_380 |
0.1061743093 |
- |
- |
PREDICTED: (-)-alpha-terpineol synthase-like [Vitis vinifera] |
| 16 |
Hb_104509_010 |
0.1073947168 |
- |
- |
photosystem II CP43 chlorophyll apoprotein (chloroplast) [Ficus sp. Moore 315] |
| 17 |
Hb_000907_010 |
0.1111549628 |
- |
- |
Ankyrin repeat-containing-like protein [Theobroma cacao] |
| 18 |
Hb_006302_050 |
0.1122366444 |
- |
- |
PREDICTED: uncharacterized protein LOC104804969, partial [Tarenaya hassleriana] |
| 19 |
Hb_106850_010 |
0.112505675 |
- |
- |
photosystem II protein D2 [Lactuca sativa] |
| 20 |
Hb_006040_020 |
0.1134826938 |
- |
- |
ATPase subunit 8 (mitochondrion) [Hevea brasiliensis] |