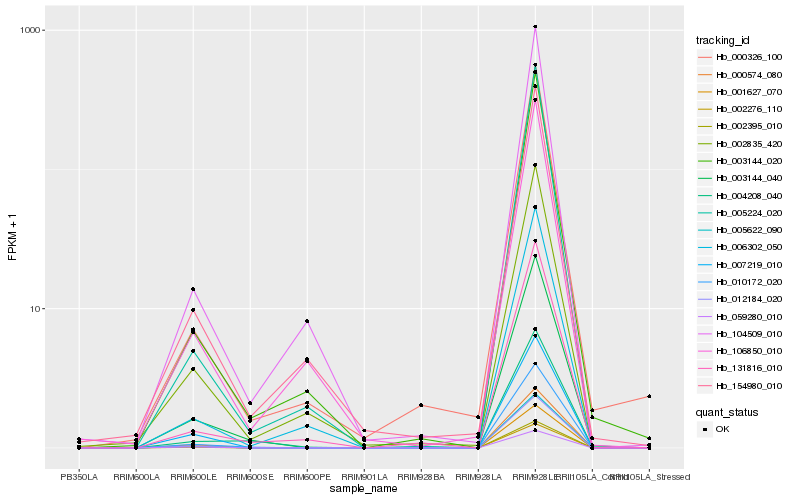
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003144_040 |
0.0 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 2 |
Hb_010172_020 |
0.0339155335 |
- |
- |
PREDICTED: tyrosine/DOPA decarboxylase 1-like [Jatropha curcas] |
| 3 |
Hb_001627_070 |
0.060584353 |
transcription factor |
TF Family: MIKC |
PREDICTED: developmental protein SEPALLATA 1 [Nelumbo nucifera] |
| 4 |
Hb_012184_020 |
0.0611827386 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000574_080 |
0.0618881456 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 6 |
Hb_002395_010 |
0.0620420212 |
- |
- |
- |
| 7 |
Hb_005622_090 |
0.0625902609 |
- |
- |
hypothetical protein CICLE_v10027074mg [Citrus clementina] |
| 8 |
Hb_003144_020 |
0.0705207047 |
- |
- |
(R)-limonene synthase, putative [Ricinus communis] |
| 9 |
Hb_007219_010 |
0.0751931156 |
- |
- |
- |
| 10 |
Hb_059280_010 |
0.0795732384 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 11 |
Hb_004208_040 |
0.0800766848 |
- |
- |
UDP-glycosyltransferase 86A1 [Morus notabilis] |
| 12 |
Hb_002835_420 |
0.0813668404 |
- |
- |
photosystem I P700 chlorophyll A apoprotein A2 [Medicago truncatula] |
| 13 |
Hb_002276_110 |
0.0814535118 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_154980_010 |
0.0821253345 |
- |
- |
photosystem I P700 chlorophyll A apoprotein A2 [Medicago truncatula] |
| 15 |
Hb_131816_010 |
0.0847170067 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 16 |
Hb_000326_100 |
0.0866908423 |
transcription factor |
TF Family: MYB-related |
DnaJ homolog subfamily C member 2 [Morus notabilis] |
| 17 |
Hb_104509_010 |
0.0870318029 |
- |
- |
photosystem II CP43 chlorophyll apoprotein (chloroplast) [Ficus sp. Moore 315] |
| 18 |
Hb_005224_020 |
0.0872581617 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_106850_010 |
0.0900643793 |
- |
- |
photosystem II protein D2 [Lactuca sativa] |
| 20 |
Hb_006302_050 |
0.0932388872 |
- |
- |
PREDICTED: uncharacterized protein LOC104804969, partial [Tarenaya hassleriana] |