| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
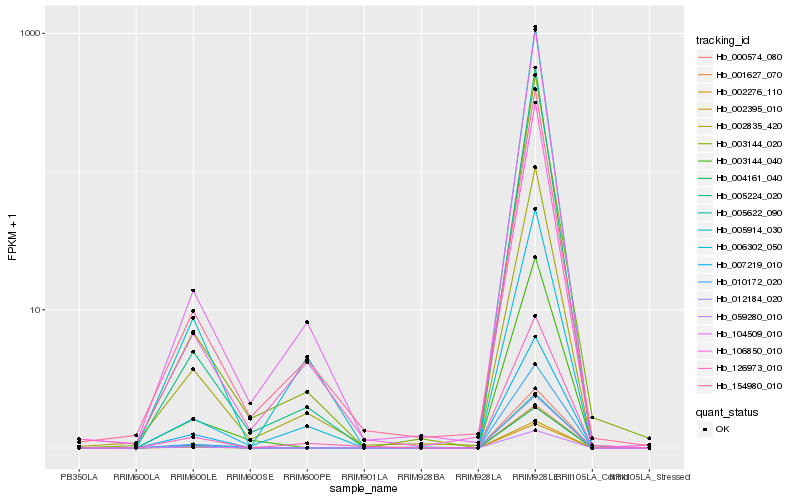
| 1 |
Hb_012184_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002395_010 |
0.0044711678 |
- |
- |
- |
| 3 |
Hb_005622_090 |
0.0067224117 |
- |
- |
hypothetical protein CICLE_v10027074mg [Citrus clementina] |
| 4 |
Hb_001627_070 |
0.0141530171 |
transcription factor |
TF Family: MIKC |
PREDICTED: developmental protein SEPALLATA 1 [Nelumbo nucifera] |
| 5 |
Hb_000574_080 |
0.0230766664 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 6 |
Hb_010172_020 |
0.0462909174 |
- |
- |
PREDICTED: tyrosine/DOPA decarboxylase 1-like [Jatropha curcas] |
| 7 |
Hb_007219_010 |
0.0543820028 |
- |
- |
- |
| 8 |
Hb_003144_040 |
0.0611827386 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 9 |
Hb_059280_010 |
0.0613840861 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 10 |
Hb_002276_110 |
0.0642208091 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_005224_020 |
0.0642420697 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_003144_020 |
0.0660597289 |
- |
- |
(R)-limonene synthase, putative [Ricinus communis] |
| 13 |
Hb_005914_030 |
0.0683227208 |
- |
- |
- |
| 14 |
Hb_104509_010 |
0.0726446365 |
- |
- |
photosystem II CP43 chlorophyll apoprotein (chloroplast) [Ficus sp. Moore 315] |
| 15 |
Hb_002835_420 |
0.0743740766 |
- |
- |
photosystem I P700 chlorophyll A apoprotein A2 [Medicago truncatula] |
| 16 |
Hb_006302_050 |
0.0756940969 |
- |
- |
PREDICTED: uncharacterized protein LOC104804969, partial [Tarenaya hassleriana] |
| 17 |
Hb_126973_010 |
0.079855748 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Vitis vinifera] |
| 18 |
Hb_004161_040 |
0.0805534261 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Populus euphratica] |
| 19 |
Hb_154980_010 |
0.0811371445 |
- |
- |
photosystem I P700 chlorophyll A apoprotein A2 [Medicago truncatula] |
| 20 |
Hb_106850_010 |
0.0813126954 |
- |
- |
photosystem II protein D2 [Lactuca sativa] |