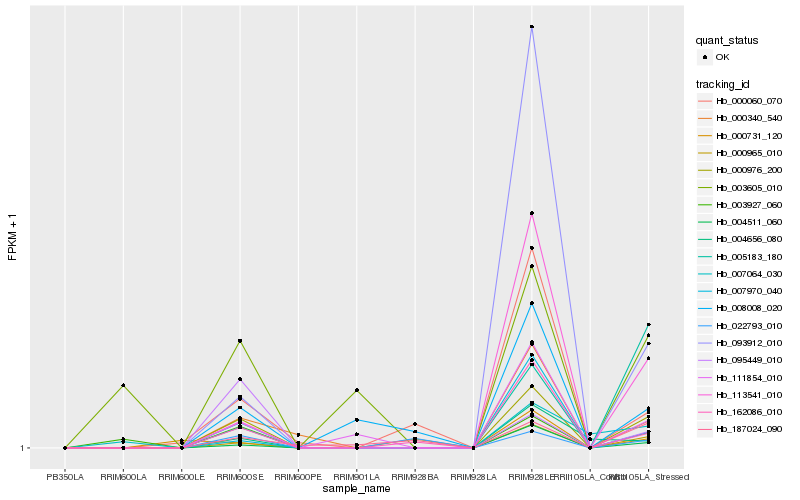
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000976_200 |
0.0 |
- |
- |
- |
| 2 |
Hb_004511_060 |
0.2054141777 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103328625 [Prunus mume] |
| 3 |
Hb_113541_010 |
0.2150191488 |
- |
- |
PREDICTED: uncharacterized protein LOC105762713 [Gossypium raimondii] |
| 4 |
Hb_000340_540 |
0.2304361075 |
- |
- |
PREDICTED: peroxidase 39-like [Jatropha curcas] |
| 5 |
Hb_111854_010 |
0.2427329334 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 6 |
Hb_162086_010 |
0.2430049465 |
- |
- |
PREDICTED: uncharacterized protein LOC105634453 [Jatropha curcas] |
| 7 |
Hb_003927_060 |
0.2501172332 |
- |
- |
hypothetical protein JCGZ_05636 [Jatropha curcas] |
| 8 |
Hb_093912_010 |
0.2511997472 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 9 |
Hb_005183_180 |
0.2764486941 |
- |
- |
alpha amylase family protein [Populus trichocarpa] |
| 10 |
Hb_007970_040 |
0.2820127686 |
- |
- |
PREDICTED: uncharacterized protein LOC104587672 [Nelumbo nucifera] |
| 11 |
Hb_000060_070 |
0.2944221598 |
- |
- |
hypothetical protein EUTSA_v10023188mg, partial [Eutrema salsugineum] |
| 12 |
Hb_095449_010 |
0.3058720739 |
- |
- |
PREDICTED: low-temperature-induced 65 kDa protein-like [Jatropha curcas] |
| 13 |
Hb_007064_030 |
0.3091257484 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 14 |
Hb_008008_020 |
0.314404357 |
- |
- |
- |
| 15 |
Hb_004656_080 |
0.3199219478 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_000731_120 |
0.322723987 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 17 |
Hb_000965_010 |
0.3313821293 |
- |
- |
hypothetical protein VITISV_016917 [Vitis vinifera] |
| 18 |
Hb_187024_090 |
0.3319291143 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_022793_010 |
0.3422000308 |
- |
- |
PREDICTED: uncharacterized protein LOC104221396 [Nicotiana sylvestris] |
| 20 |
Hb_003605_010 |
0.347314385 |
- |
- |
- |