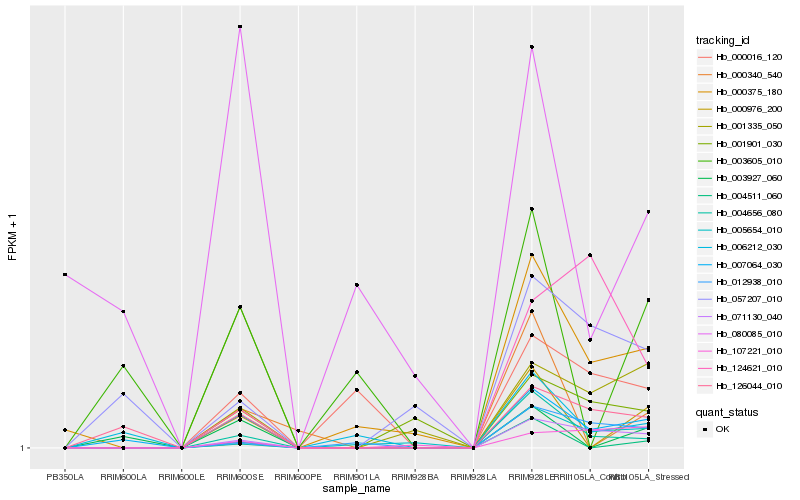
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007064_030 |
0.0 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 2 |
Hb_126044_010 |
0.1436358023 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 3 |
Hb_071130_040 |
0.2411806156 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004656_080 |
0.2519374691 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_003927_060 |
0.2656044187 |
- |
- |
hypothetical protein JCGZ_05636 [Jatropha curcas] |
| 6 |
Hb_057207_010 |
0.2838911764 |
- |
- |
hypothetical protein F383_17010 [Gossypium arboreum] |
| 7 |
Hb_001335_050 |
0.2902068061 |
- |
- |
- |
| 8 |
Hb_000976_200 |
0.3091257484 |
- |
- |
- |
| 9 |
Hb_006212_030 |
0.324483586 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 10 |
Hb_005654_010 |
0.3326942491 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_012938_010 |
0.3417427359 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 12 |
Hb_000375_180 |
0.3442727149 |
- |
- |
PREDICTED: uncharacterized protein LOC104593656 [Nelumbo nucifera] |
| 13 |
Hb_107221_010 |
0.3445468346 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_124621_010 |
0.3504361319 |
- |
- |
hypothetical protein L484_019698 [Morus notabilis] |
| 15 |
Hb_003605_010 |
0.3511282256 |
- |
- |
- |
| 16 |
Hb_000016_120 |
0.3524837668 |
- |
- |
PREDICTED: uncharacterized protein LOC104610208 [Nelumbo nucifera] |
| 17 |
Hb_001901_030 |
0.3562929642 |
- |
- |
- |
| 18 |
Hb_080085_010 |
0.3726881101 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 19 |
Hb_004511_060 |
0.3765254953 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103328625 [Prunus mume] |
| 20 |
Hb_000340_540 |
0.3792856727 |
- |
- |
PREDICTED: peroxidase 39-like [Jatropha curcas] |