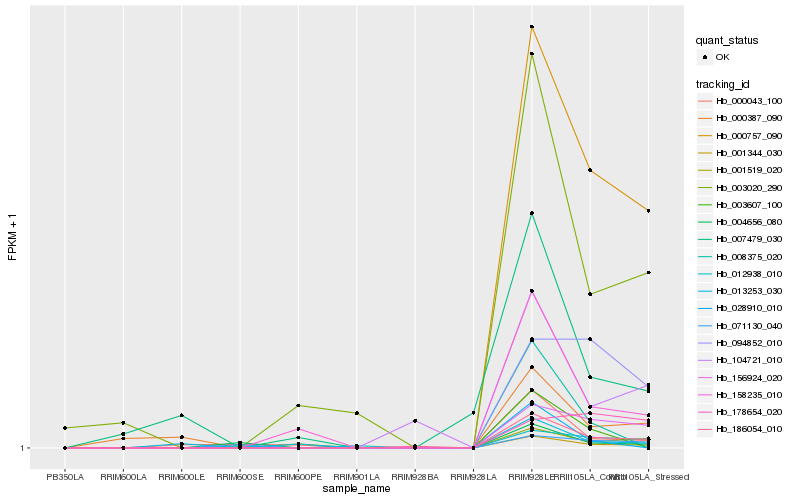
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001344_030 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_000757_090 |
0.1219191442 |
- |
- |
SPla/RYanodine receptor domain-containing protein isoform 3 [Theobroma cacao] |
| 3 |
Hb_186054_010 |
0.1219887504 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 4 |
Hb_156924_020 |
0.1616649569 |
- |
- |
PREDICTED: uncharacterized protein LOC105644346 [Jatropha curcas] |
| 5 |
Hb_158235_010 |
0.1933139457 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 6 |
Hb_104721_010 |
0.2256237896 |
- |
- |
PREDICTED: uncharacterized protein LOC105763691 [Gossypium raimondii] |
| 7 |
Hb_094852_010 |
0.2273769901 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 8 |
Hb_003020_290 |
0.2373380367 |
- |
- |
hypothetical protein CICLE_v10029104mg [Citrus clementina] |
| 9 |
Hb_001519_020 |
0.2562291854 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 10 |
Hb_000387_090 |
0.2652907879 |
- |
- |
PREDICTED: uncharacterized protein LOC104115978 [Nicotiana tomentosiformis] |
| 11 |
Hb_000043_100 |
0.2677013234 |
- |
- |
PREDICTED: uncharacterized protein LOC105797583 [Gossypium raimondii] |
| 12 |
Hb_012938_010 |
0.2686365531 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 13 |
Hb_004656_080 |
0.2793489395 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_071130_040 |
0.2809296643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003607_100 |
0.2842682157 |
- |
- |
PREDICTED: zinc finger protein 3 [Jatropha curcas] |
| 16 |
Hb_008375_020 |
0.2892864627 |
- |
- |
PREDICTED: uncharacterized protein LOC104894372 [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_178654_020 |
0.2945793034 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 18 |
Hb_013253_030 |
0.3013715319 |
- |
- |
PREDICTED: protein argonaute 16 isoform X1 [Jatropha curcas] |
| 19 |
Hb_028910_010 |
0.3028879543 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 20 |
Hb_007479_030 |
0.3041618829 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |