| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
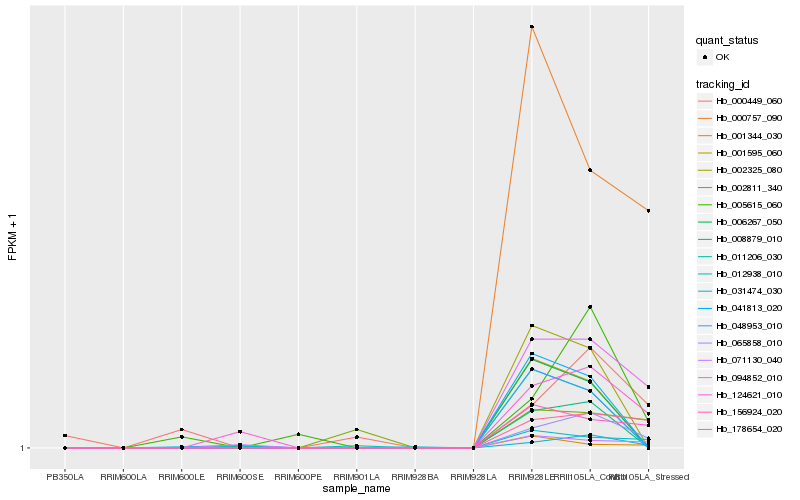
Hb_065858_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104227769, partial [Nicotiana sylvestris] |
| 2 |
Hb_094852_010 |
0.1339806436 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 3 |
Hb_178654_020 |
0.1773538594 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 4 |
Hb_124621_010 |
0.2139004751 |
- |
- |
hypothetical protein L484_019698 [Morus notabilis] |
| 5 |
Hb_156924_020 |
0.2191694776 |
- |
- |
PREDICTED: uncharacterized protein LOC105644346 [Jatropha curcas] |
| 6 |
Hb_005615_060 |
0.2439129383 |
- |
- |
PREDICTED: uncharacterized protein LOC105636752 [Jatropha curcas] |
| 7 |
Hb_000757_090 |
0.2464886849 |
- |
- |
SPla/RYanodine receptor domain-containing protein isoform 3 [Theobroma cacao] |
| 8 |
Hb_008879_010 |
0.286160652 |
- |
- |
Beta-ureidopropionase, putative [Ricinus communis] |
| 9 |
Hb_031474_030 |
0.3111176739 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 10 |
Hb_000449_060 |
0.3178623794 |
- |
- |
- |
| 11 |
Hb_012938_010 |
0.3186199552 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 12 |
Hb_002325_080 |
0.3240824242 |
- |
- |
- |
| 13 |
Hb_071130_040 |
0.327477397 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_041813_020 |
0.3319494258 |
- |
- |
ATP synthase epsilon subunit [Grubbia rosmarinifolia] |
| 15 |
Hb_001595_060 |
0.3334034077 |
- |
- |
hypothetical protein L484_001429 [Morus notabilis] |
| 16 |
Hb_006267_050 |
0.3336776648 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 17 |
Hb_002811_340 |
0.3345399441 |
- |
- |
PREDICTED: ammonium transporter 3 member 2-like [Populus euphratica] |
| 18 |
Hb_001344_030 |
0.3357883601 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_011206_030 |
0.3365924709 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit Rieske-4, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_048953_010 |
0.3365924709 |
- |
- |
PREDICTED: uncharacterized protein LOC105775428 [Gossypium raimondii] |