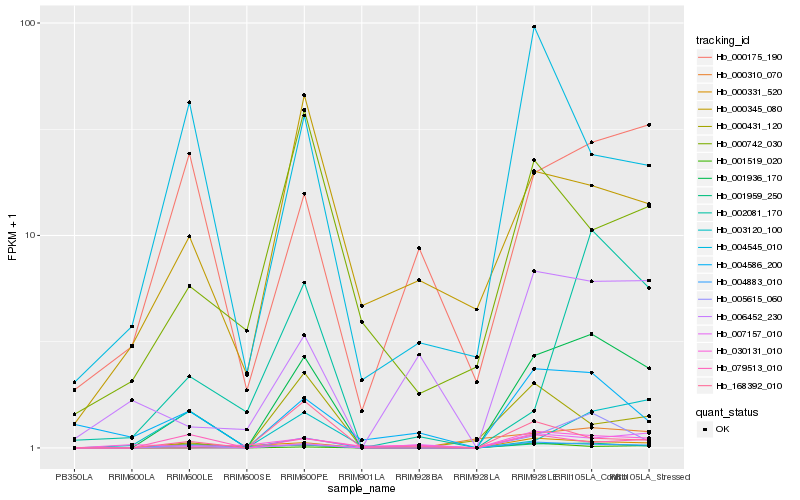
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001936_170 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004883_010 |
0.2295683693 |
- |
- |
hypothetical protein JCGZ_23144 [Jatropha curcas] |
| 3 |
Hb_001959_250 |
0.2580828458 |
- |
- |
hypothetical protein CARUB_v10015435mg [Capsella rubella] |
| 4 |
Hb_005615_060 |
0.2984703806 |
- |
- |
PREDICTED: uncharacterized protein LOC105636752 [Jatropha curcas] |
| 5 |
Hb_002081_170 |
0.3098366723 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 6 |
Hb_000431_120 |
0.3144472396 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1-like [Jatropha curcas] |
| 7 |
Hb_006452_230 |
0.3166569117 |
- |
- |
PREDICTED: uncharacterized protein LOC105635044 [Jatropha curcas] |
| 8 |
Hb_004586_200 |
0.3172249493 |
- |
- |
PREDICTED: cyclin-D3-1-like [Jatropha curcas] |
| 9 |
Hb_000175_190 |
0.3201092764 |
- |
- |
PREDICTED: phosphatidylcholine:diacylglycerol cholinephosphotransferase 1-like [Jatropha curcas] |
| 10 |
Hb_030131_010 |
0.3251342994 |
- |
- |
PREDICTED: probable acyl-activating enzyme 1, peroxisomal, partial [Jatropha curcas] |
| 11 |
Hb_000742_030 |
0.3345738148 |
- |
- |
PREDICTED: uncharacterized protein LOC105636851 [Jatropha curcas] |
| 12 |
Hb_001519_020 |
0.3378392467 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 13 |
Hb_168392_010 |
0.3379608831 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 14 |
Hb_079513_010 |
0.3381398211 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_007157_010 |
0.3385657374 |
- |
- |
- |
| 16 |
Hb_003120_100 |
0.341780577 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 17 |
Hb_000345_080 |
0.3442157598 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 18 |
Hb_000331_520 |
0.3465704266 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 19 |
Hb_004545_010 |
0.3483759583 |
- |
- |
acyl carrier protein [Ricinus communis] |
| 20 |
Hb_000310_070 |
0.349978103 |
- |
- |
BnaA06g08330D [Brassica napus] |