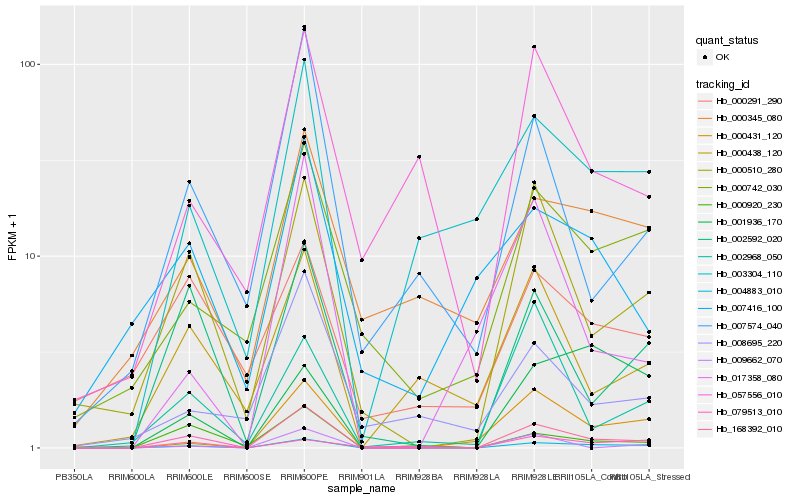
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004883_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_23144 [Jatropha curcas] |
| 2 |
Hb_168392_010 |
0.1315696115 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 3 |
Hb_000431_120 |
0.2088551024 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_000510_280 |
0.2276059612 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-induced protein 22D-like [Jatropha curcas] |
| 5 |
Hb_001936_170 |
0.2295683693 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000742_030 |
0.2379651457 |
- |
- |
PREDICTED: uncharacterized protein LOC105636851 [Jatropha curcas] |
| 7 |
Hb_003304_110 |
0.2441214185 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002592_020 |
0.2442809468 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000345_080 |
0.2709297634 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 10 |
Hb_000920_230 |
0.2728088513 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_000438_120 |
0.2951608812 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 12 |
Hb_002968_050 |
0.2980608945 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 13 |
Hb_057556_010 |
0.3047242342 |
- |
- |
Light-regulated protein precursor, putative [Ricinus communis] |
| 14 |
Hb_007416_100 |
0.308123624 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 5 [Jatropha curcas] |
| 15 |
Hb_007574_040 |
0.3082681642 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |
| 16 |
Hb_017358_080 |
0.3083476562 |
- |
- |
- |
| 17 |
Hb_009662_070 |
0.3084577392 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 18 |
Hb_008695_220 |
0.3091862119 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 19 |
Hb_079513_010 |
0.3098700866 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000291_290 |
0.310897445 |
- |
- |
conserved hypothetical protein [Ricinus communis] |