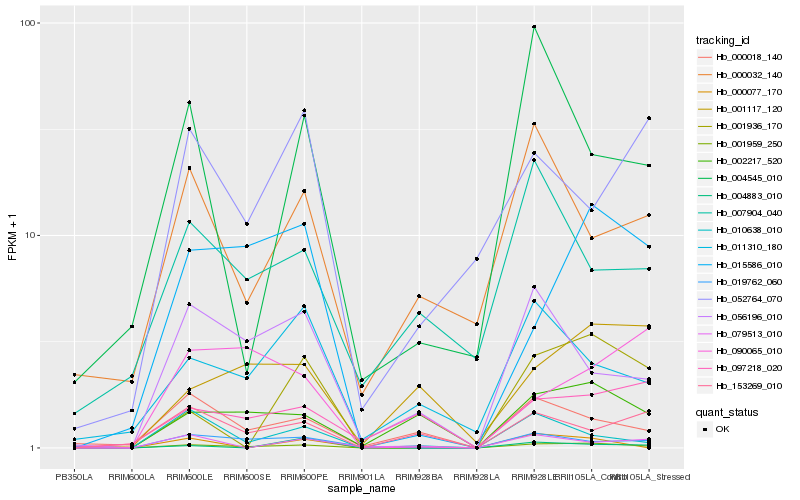
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001959_250 |
0.0 |
- |
- |
hypothetical protein CARUB_v10015435mg [Capsella rubella] |
| 2 |
Hb_019762_060 |
0.2431152439 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_015586_010 |
0.2497190548 |
- |
- |
PREDICTED: flavonol sulfotransferase-like [Jatropha curcas] |
| 4 |
Hb_001936_170 |
0.2580828458 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002217_520 |
0.2630611375 |
- |
- |
PREDICTED: probable protein phosphatase 2C 42 isoform X3 [Populus euphratica] |
| 6 |
Hb_056196_010 |
0.2701904411 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 7 |
Hb_010638_010 |
0.2765215619 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigC [Jatropha curcas] |
| 8 |
Hb_000018_140 |
0.2792953263 |
- |
- |
PREDICTED: uncharacterized protein LOC105638239 [Jatropha curcas] |
| 9 |
Hb_011310_180 |
0.2885460378 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_004545_010 |
0.3060043509 |
- |
- |
acyl carrier protein [Ricinus communis] |
| 11 |
Hb_153269_010 |
0.3069616536 |
- |
- |
hypothetical protein CICLE_v10029937mg [Citrus clementina] |
| 12 |
Hb_000032_140 |
0.3084947568 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_090065_010 |
0.3123382651 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 14 |
Hb_001117_120 |
0.3132792643 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 15 |
Hb_004883_010 |
0.3138749741 |
- |
- |
hypothetical protein JCGZ_23144 [Jatropha curcas] |
| 16 |
Hb_052764_070 |
0.3148956216 |
- |
- |
PREDICTED: uncharacterized protein LOC105649473 [Jatropha curcas] |
| 17 |
Hb_097218_020 |
0.3156301824 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit E [Jatropha curcas] |
| 18 |
Hb_079513_010 |
0.3159080659 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_007904_040 |
0.3191169287 |
- |
- |
PREDICTED: uncharacterized protein LOC105644727 [Jatropha curcas] |
| 20 |
Hb_000077_170 |
0.3201519452 |
- |
- |
PREDICTED: GPI-anchored protein LORELEI-like [Jatropha curcas] |