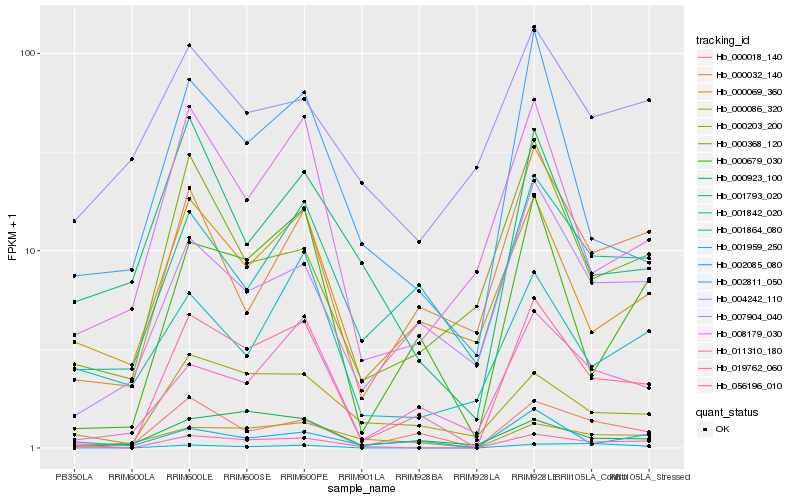
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019762_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_056196_010 |
0.1826701972 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 3 |
Hb_000203_200 |
0.2170105698 |
- |
- |
Pollen allergen Che a 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_000086_320 |
0.2211913457 |
- |
- |
hypothetical protein JCGZ_17661 [Jatropha curcas] |
| 5 |
Hb_000018_140 |
0.2394375221 |
- |
- |
PREDICTED: uncharacterized protein LOC105638239 [Jatropha curcas] |
| 6 |
Hb_001959_250 |
0.2431152439 |
- |
- |
hypothetical protein CARUB_v10015435mg [Capsella rubella] |
| 7 |
Hb_000679_030 |
0.2529975321 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HY5 [Jatropha curcas] |
| 8 |
Hb_001842_020 |
0.2530791927 |
- |
- |
PREDICTED: low molecular weight phosphotyrosine protein phosphatase [Jatropha curcas] |
| 9 |
Hb_001793_020 |
0.2547123329 |
- |
- |
PREDICTED: uncharacterized protein LOC105628624 [Jatropha curcas] |
| 10 |
Hb_008179_030 |
0.2575679627 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 8 [Jatropha curcas] |
| 11 |
Hb_000368_120 |
0.2594184764 |
- |
- |
PREDICTED: uncharacterized protein LOC105633862 [Jatropha curcas] |
| 12 |
Hb_011310_180 |
0.2597148862 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_007904_040 |
0.2597520249 |
- |
- |
PREDICTED: uncharacterized protein LOC105644727 [Jatropha curcas] |
| 14 |
Hb_000069_360 |
0.2602922251 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004242_110 |
0.2614285293 |
- |
- |
PREDICTED: uncharacterized protein LOC105632860 [Jatropha curcas] |
| 16 |
Hb_000032_140 |
0.2625972875 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000923_100 |
0.2635317494 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 18 |
Hb_001864_080 |
0.26373783 |
- |
- |
PREDICTED: uncharacterized protein LOC105649405 [Jatropha curcas] |
| 19 |
Hb_002085_080 |
0.267496477 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 68-like [Jatropha curcas] |
| 20 |
Hb_002811_050 |
0.2677722391 |
- |
- |
PREDICTED: uncharacterized protein LOC105647652 [Jatropha curcas] |