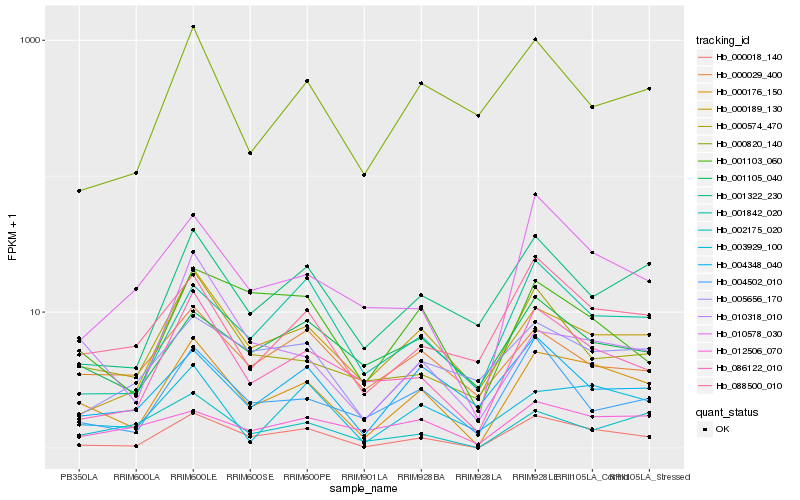
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000176_150 |
0.0 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000018_140 |
0.1338414843 |
- |
- |
PREDICTED: uncharacterized protein LOC105638239 [Jatropha curcas] |
| 3 |
Hb_086122_010 |
0.1758784372 |
- |
- |
PREDICTED: immunoglobulin-like domain-containing receptor 1 [Jatropha curcas] |
| 4 |
Hb_003929_100 |
0.1842545064 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL61 [Populus euphratica] |
| 5 |
Hb_088500_010 |
0.1888681808 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At4g39970 [Jatropha curcas] |
| 6 |
Hb_000189_130 |
0.1906274311 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATA, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001322_230 |
0.2022914714 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 8 |
Hb_010578_030 |
0.2054975943 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATA, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001103_060 |
0.2069889467 |
- |
- |
proton-dependent oligopeptide transport family protein [Populus trichocarpa] |
| 10 |
Hb_002175_020 |
0.2095724069 |
- |
- |
PREDICTED: uncharacterized protein LOC105642125 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001842_020 |
0.2105508831 |
- |
- |
PREDICTED: low molecular weight phosphotyrosine protein phosphatase [Jatropha curcas] |
| 12 |
Hb_004348_040 |
0.2124922939 |
- |
- |
type I inositol polyphosphate 5-phosphatase, putative [Ricinus communis] |
| 13 |
Hb_001105_040 |
0.2125290242 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 10, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_004502_010 |
0.2132153435 |
- |
- |
hypothetical protein JCGZ_23576 [Jatropha curcas] |
| 15 |
Hb_012506_070 |
0.2138100415 |
- |
- |
hypothetical protein POPTR_0004s20060g [Populus trichocarpa] |
| 16 |
Hb_000574_470 |
0.2168150977 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_005656_170 |
0.2186094119 |
- |
- |
PREDICTED: uncharacterized protein LOC105637447 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000029_400 |
0.2188209336 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |
| 19 |
Hb_010318_010 |
0.2196655268 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000820_140 |
0.2209266966 |
- |
- |
histone H4 [Zea mays] |