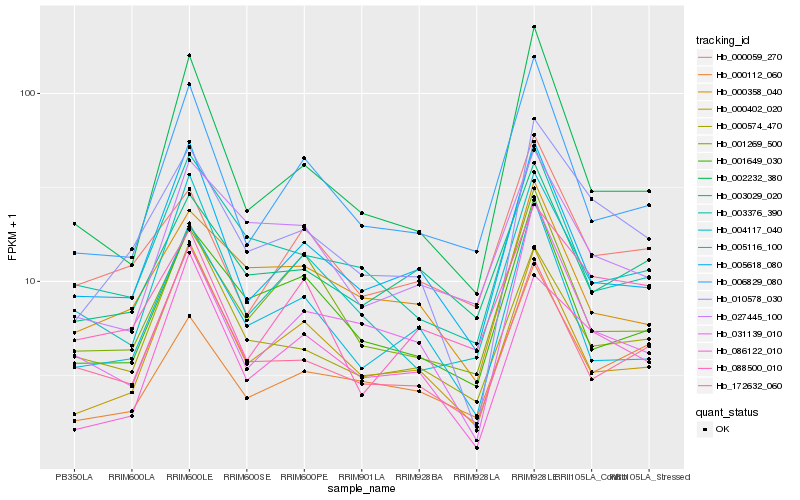
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010578_030 |
0.0 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATA, chloroplastic [Jatropha curcas] |
| 2 |
Hb_086122_010 |
0.1427208554 |
- |
- |
PREDICTED: immunoglobulin-like domain-containing receptor 1 [Jatropha curcas] |
| 3 |
Hb_031139_010 |
0.14938589 |
- |
- |
PREDICTED: uncharacterized protein LOC105632403 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000358_040 |
0.1538112991 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 4, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000112_060 |
0.1559410767 |
- |
- |
PREDICTED: thioredoxin-like protein CITRX, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000402_020 |
0.1580254868 |
- |
- |
PREDICTED: 2-phytyl-1,4-beta-naphthoquinone methyltransferase, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_000059_270 |
0.1614941189 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 8 |
Hb_005618_080 |
0.1621400619 |
- |
- |
PREDICTED: protoporphyrinogen oxidase 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_004117_040 |
0.1632866548 |
- |
- |
PREDICTED: uncharacterized protein LOC105638287 [Jatropha curcas] |
| 10 |
Hb_088500_010 |
0.1639730069 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At4g39970 [Jatropha curcas] |
| 11 |
Hb_005116_100 |
0.1644705793 |
- |
- |
PREDICTED: uncharacterized protein LOC105649999 [Jatropha curcas] |
| 12 |
Hb_001649_030 |
0.1678371381 |
- |
- |
PREDICTED: peptide deformylase 1B, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003029_020 |
0.1683801653 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 14 |
Hb_002232_380 |
0.1685864258 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_003376_390 |
0.1697300765 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001269_500 |
0.1717660844 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 17 |
Hb_000574_470 |
0.1742423722 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X1 [Jatropha curcas] |
| 18 |
Hb_172632_060 |
0.1755716878 |
- |
- |
PREDICTED: uncharacterized protein LOC105646135 isoform X1 [Jatropha curcas] |
| 19 |
Hb_006829_080 |
0.1760572296 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 20 |
Hb_027445_100 |
0.1764046702 |
- |
- |
PREDICTED: probable monodehydroascorbate reductase, cytoplasmic isoform 2 [Jatropha curcas] |