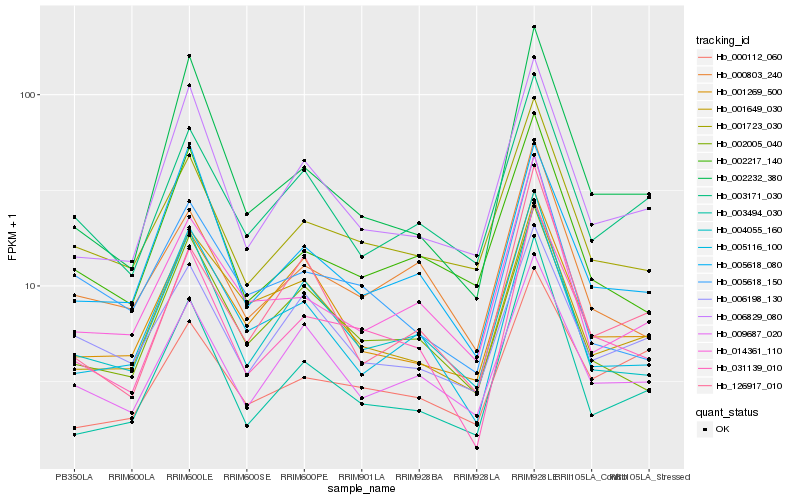
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031139_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632403 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002232_380 |
0.1094925816 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_002005_040 |
0.1160325182 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 4 |
Hb_004055_160 |
0.1164649275 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_006829_080 |
0.1194619383 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 6 |
Hb_009687_020 |
0.1197914239 |
- |
- |
hypothetical protein JCGZ_10323 [Jatropha curcas] |
| 7 |
Hb_000803_240 |
0.121359634 |
- |
- |
hypothetical protein CICLE_v100143612mg, partial [Citrus clementina] |
| 8 |
Hb_005618_080 |
0.1229806026 |
- |
- |
PREDICTED: protoporphyrinogen oxidase 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001723_030 |
0.1249573372 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_005116_100 |
0.1259110344 |
- |
- |
PREDICTED: uncharacterized protein LOC105649999 [Jatropha curcas] |
| 11 |
Hb_002217_140 |
0.1316025696 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Nelumbo nucifera] |
| 12 |
Hb_014361_110 |
0.1353939165 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_003171_030 |
0.1395656736 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 14 |
Hb_001269_500 |
0.1404966621 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 15 |
Hb_005618_150 |
0.1405034051 |
- |
- |
PREDICTED: protein translocase subunit SecA, chloroplastic [Jatropha curcas] |
| 16 |
Hb_003494_030 |
0.1428273926 |
- |
- |
PREDICTED: protease Do-like 8, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001649_030 |
0.1431783071 |
- |
- |
PREDICTED: peptide deformylase 1B, chloroplastic [Jatropha curcas] |
| 18 |
Hb_006198_130 |
0.1432638394 |
- |
- |
PREDICTED: uncharacterized protein LOC105644406 [Jatropha curcas] |
| 19 |
Hb_000112_060 |
0.1439529524 |
- |
- |
PREDICTED: thioredoxin-like protein CITRX, chloroplastic [Jatropha curcas] |
| 20 |
Hb_126917_010 |
0.1441927581 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |