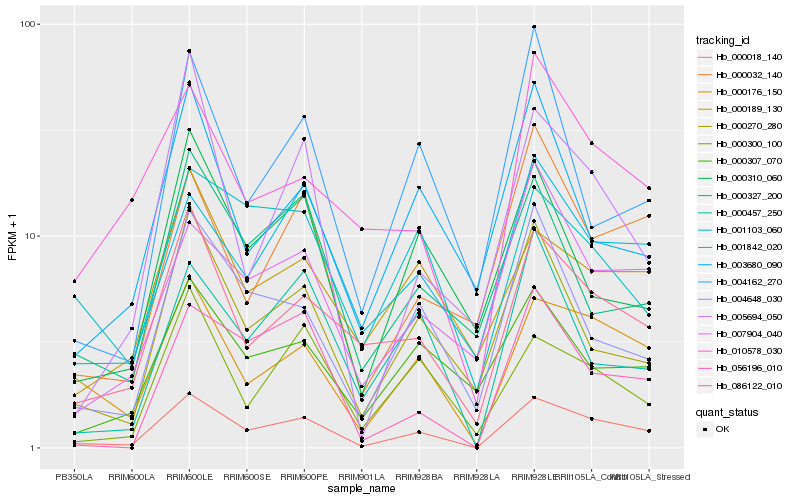
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000018_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638239 [Jatropha curcas] |
| 2 |
Hb_000176_150 |
0.1338414843 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_086122_010 |
0.1559348989 |
- |
- |
PREDICTED: immunoglobulin-like domain-containing receptor 1 [Jatropha curcas] |
| 4 |
Hb_000270_280 |
0.1735242216 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 5 |
Hb_004648_030 |
0.1742383671 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 6 |
Hb_001842_020 |
0.1775649872 |
- |
- |
PREDICTED: low molecular weight phosphotyrosine protein phosphatase [Jatropha curcas] |
| 7 |
Hb_056196_010 |
0.1833239803 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 8 |
Hb_000300_100 |
0.1836997763 |
- |
- |
NADH-cytochrome b5 reductase 1 [Morus notabilis] |
| 9 |
Hb_007904_040 |
0.186067501 |
- |
- |
PREDICTED: uncharacterized protein LOC105644727 [Jatropha curcas] |
| 10 |
Hb_000032_140 |
0.1861778534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000457_250 |
0.1875405642 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 12 |
Hb_000307_070 |
0.1910300233 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 13 |
Hb_000310_060 |
0.1928259403 |
- |
- |
hypothetical protein JCGZ_20793 [Jatropha curcas] |
| 14 |
Hb_000189_130 |
0.1952203462 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATA, chloroplastic [Jatropha curcas] |
| 15 |
Hb_010578_030 |
0.1956067572 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATA, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001103_060 |
0.198457617 |
- |
- |
proton-dependent oligopeptide transport family protein [Populus trichocarpa] |
| 17 |
Hb_000327_200 |
0.1988146456 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 18 |
Hb_005694_050 |
0.1988541867 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 19 |
Hb_004162_270 |
0.2010793914 |
- |
- |
PREDICTED: superoxide dismutase [Cu-Zn], chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_003680_090 |
0.2031907841 |
- |
- |
PREDICTED: threonine dehydratase biosynthetic, chloroplastic [Jatropha curcas] |