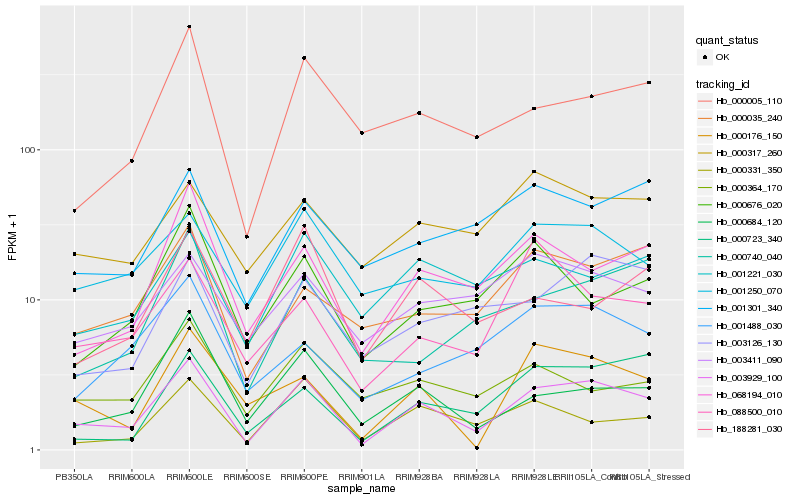
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003929_100 |
0.0 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL61 [Populus euphratica] |
| 2 |
Hb_068194_010 |
0.1709187309 |
- |
- |
hypothetical protein POPTR_0010s23740g [Populus trichocarpa] |
| 3 |
Hb_000005_110 |
0.1816309408 |
- |
- |
histone H2B1 [Hevea brasiliensis] |
| 4 |
Hb_000331_350 |
0.1837995752 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 5 |
Hb_000176_150 |
0.1842545064 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001250_070 |
0.1858595813 |
- |
- |
ATP-citrate synthase, putative [Ricinus communis] |
| 7 |
Hb_003126_130 |
0.1882194024 |
- |
- |
inositol-1-monophosphatase family protein [Populus trichocarpa] |
| 8 |
Hb_000684_120 |
0.1887062996 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000723_340 |
0.1924041572 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG3 [Jatropha curcas] |
| 10 |
Hb_001221_030 |
0.1925273636 |
- |
- |
PREDICTED: adenylate kinase 4 [Jatropha curcas] |
| 11 |
Hb_000035_240 |
0.1931359351 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001301_340 |
0.1976886619 |
- |
- |
endoribonuclease L-PSP family protein [Populus trichocarpa] |
| 13 |
Hb_000364_170 |
0.1977198716 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 14 |
Hb_000676_020 |
0.1981609612 |
- |
- |
PREDICTED: uncharacterized protein LOC105645386 [Jatropha curcas] |
| 15 |
Hb_001488_030 |
0.1985298865 |
- |
- |
biotin carboxyl carrier protein [Vernicia fordii] |
| 16 |
Hb_003411_090 |
0.198812811 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 4 of pyruvate dehydrogenase complex |
Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_000740_040 |
0.198978074 |
- |
- |
PREDICTED: uncharacterized protein LOC105126082 [Populus euphratica] |
| 18 |
Hb_000317_260 |
0.20096664 |
- |
- |
unknown [Populus trichocarpa] |
| 19 |
Hb_188281_030 |
0.2013764116 |
- |
- |
PREDICTED: uridine nucleosidase 1 [Jatropha curcas] |
| 20 |
Hb_088500_010 |
0.2030588921 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At4g39970 [Jatropha curcas] |